Author: Denis Avetisyan
Researchers have developed a powerful new method for analyzing EEG data, enabling a deeper understanding of brain connectivity and its alterations in neurological conditions.
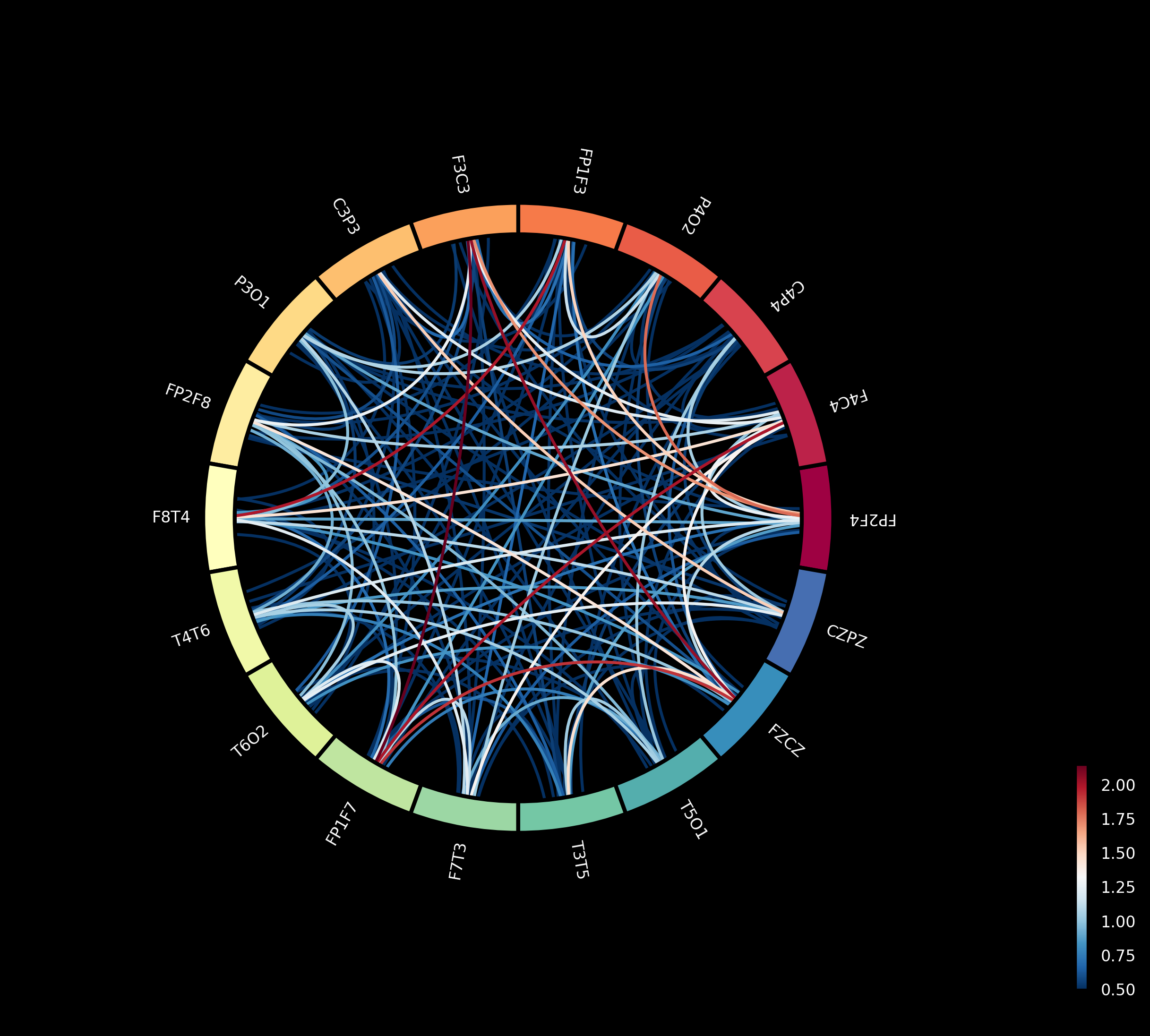
This paper introduces a novel network of conditionally coupled dynamic causal models (NccDCM) for characterizing brain connectivity and identifying disease-related patterns in epilepsy using EEG and mixed-effects modeling.
Characterizing dynamic brain connectivity across groups remains challenging due to the complexity of modeling large-scale neural interactions. This limitation is addressed in ‘Differential Dynamic Causal Nets: Model Construction, Identification and Group Comparisons’ which introduces a novel network of conditionally coupled dynamic causal models (NccDCM) for analyzing electroencephalogram (EEG) data. The proposed method infers network parameters using a loss function derived from stochastic differential equations and demonstrates its efficacy on both synthetic and real EEG data, revealing disruptions in epileptic brain connectivity. Can this approach ultimately provide a more nuanced understanding of network dynamics and improve diagnostic capabilities for neurological disorders?
Unraveling the Epileptic Network: Beyond the Local Spark
Epilepsy is increasingly understood not simply as localized neuronal misfiring, but as a systemic disruption of intricate brain networks. Traditional diagnostic tools, like electroencephalograms (EEGs), often focus on identifying seizure origins, yet struggle to capture the widespread, dynamic changes occurring within these networks that predispose an individual to seizures. The brain’s functional architecture – the coordinated activity between regions responsible for everything from cognition to motor control – becomes destabilized, leading to hypersynchrony and the characteristic clinical manifestations of epilepsy. This network dysfunction manifests as altered communication patterns, reduced network efficiency, and the formation of aberrant connections, presenting a significant challenge for accurately diagnosing and treating the condition, as the source of the problem extends beyond a single, identifiable location.
The brain doesn’t experience epileptic seizures as isolated events within a single region; rather, they emerge from the shifting and often chaotic interactions between distributed neural networks. Investigation into the preictal period – the time leading up to a seizure – reveals subtle alterations in network connectivity and activity patterns, offering a potential window for identifying predictive biomarkers. Simultaneously, characterizing the network dynamics during an ictal event – the seizure itself – is crucial for understanding how dysfunction propagates and amplifies, paving the way for targeted therapeutic interventions. Advanced neuroimaging techniques and computational modeling are increasingly employed to map these dynamic shifts, with the ultimate goal of not only diagnosing epilepsy more accurately but also anticipating seizures and developing personalized treatments that disrupt pathological network activity without impacting healthy brain function.
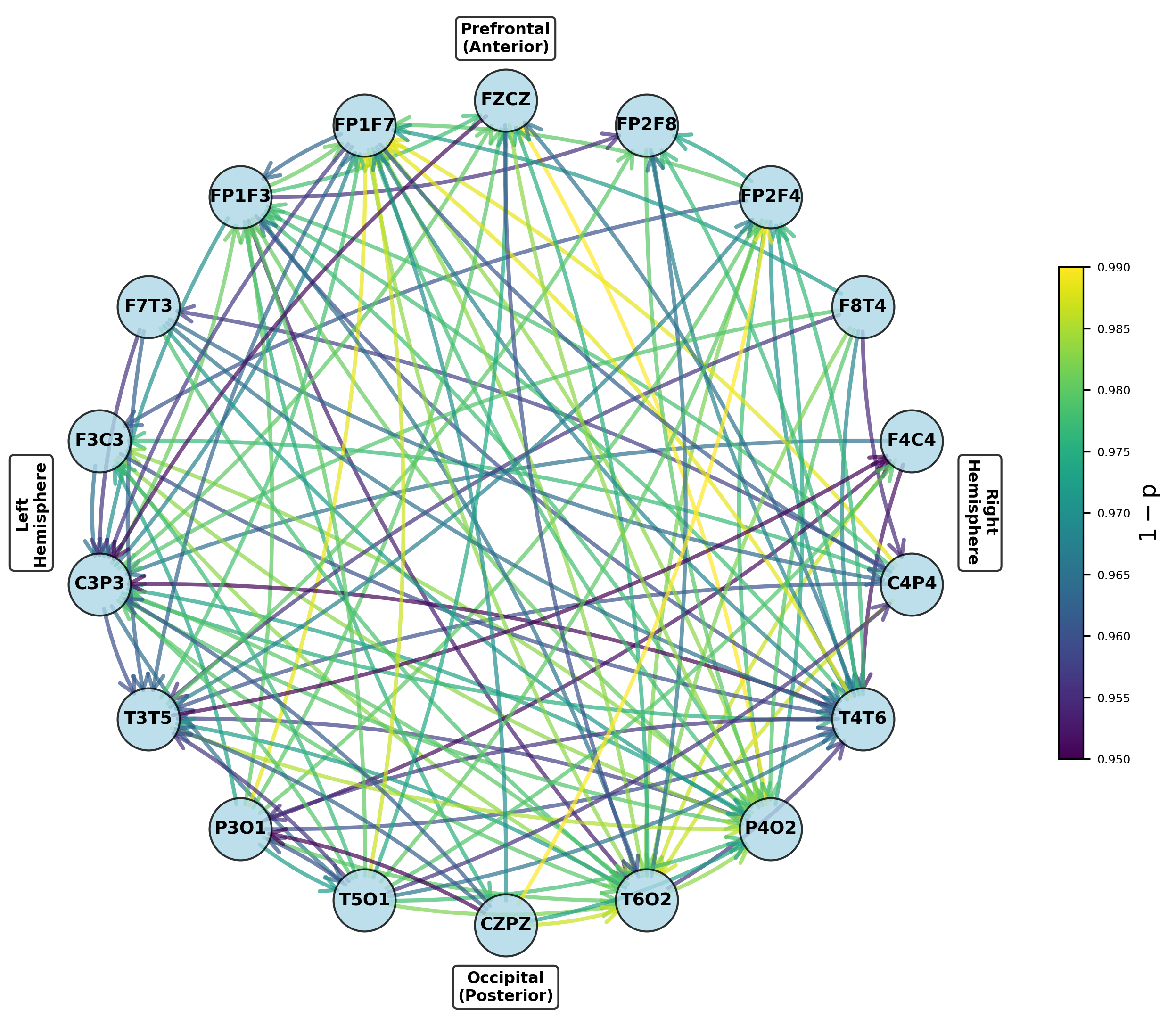
Modeling the Brain as a Network: The NccDCM Approach
The Neural Connectivity Dynamic Causal Model (NccDCM) employs conditionally coupled Dynamic Causal Models (ccDCMs) as its core components to investigate brain-wide interactions. These ccDCMs are not treated as independent units; instead, they are interconnected based on conditional coupling, meaning the influence of one ccDCM on another is determined by the activity states of both. This approach allows the NccDCM to move beyond modeling isolated brain regions and instead represent the brain as a dynamic network where activity in one area directly modulates the behavior of others. The conditional coupling scheme enables investigation of how disruptions in these connections manifest during neurological events, such as epilepsy, and how these disruptions impact overall brain function.
The Jansen-Rit model, forming the basis of each conditionally coupled Dynamic Causal Model (ccDCM) within the NccDCM framework, represents brain activity via interconnected neural populations. Specifically, it utilizes a system of four coupled differential equations to model the average postsynaptic potentials within distinct neuronal populations – pyramidal cells and two types of inhibitory interneurons. These equations detail the dynamics of excitatory and inhibitory postsynaptic currents, incorporating parameters that represent synaptic coupling strengths, time constants for synaptic transmission, and the relative proportions of each neuronal population. This approach allows the ccDCM to simulate the macroscopic electrical activity resulting from complex synaptic interactions, providing a biologically plausible representation of neural dynamics at a network level.
The NccDCM facilitates network-level analysis of brain activity by establishing connections between individual conditionally coupled Dynamic Causal Models (ccDCMs). This interconnected framework allows researchers to model the propagation of neural activity across brain regions and, crucially, to pinpoint disruptions in these connections that occur during epileptic events. By observing alterations in the modeled signal transmission between ccDCMs, the NccDCM can identify regions exhibiting abnormal connectivity or reduced signal propagation, potentially localizing the epileptogenic zone and characterizing the spread of seizure activity. This approach moves beyond single-region analysis to provide a comprehensive view of network dysfunction in epilepsy.
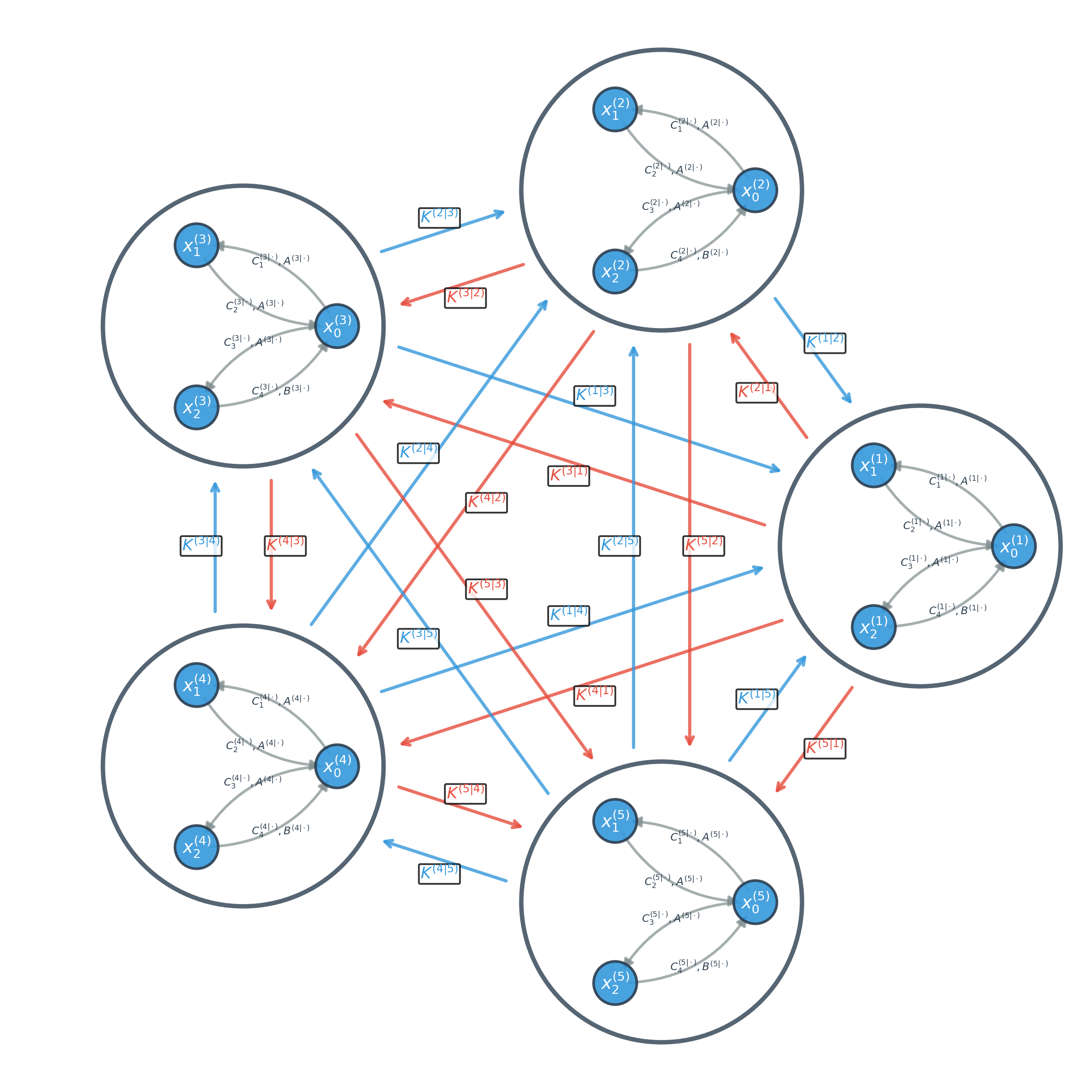
Refining the Model: From Estimation to Simplification
The NccDCM’s efficacy relies on precise parameter estimation, which is achieved through application of the Chen-Fliess Expansion. This mathematical technique provides a solution to the stochastic differential equations that define the model’s underlying dynamics. Specifically, the Chen-Fliess Expansion allows for the approximation of the transition probability density function of the system, enabling the calculation of parameters that best fit observed data. This process effectively transforms the continuous-time stochastic equations into a series of algebraic equations that can be solved numerically, yielding estimates for the model’s parameters and ensuring accurate representation of neural processes.
The Evolutionary Optimization Algorithm (EOA) is implemented to refine parameters initially estimated via the Chen-Fliess Expansion, thereby improving the NccDCM’s ability to model brain dynamics. EOA functions as a stochastic global optimization technique, iteratively evolving a population of parameter sets through processes analogous to natural selection – including mutation, crossover, and selection – based on a fitness function that quantifies model performance against empirical data. This process minimizes the discrepancy between simulated and observed brain activity, resulting in a more accurate and robust parameter configuration. The algorithm continues iterating until a pre-defined convergence criterion is met, ensuring the identified parameters represent a statistically significant and reliable representation of the underlying neural processes.
Marginal Screening techniques, applied within the NccDCM framework, function by evaluating the contribution of each connection – represented as a parameter within the model – to the overall model evidence. This is typically achieved through Bayesian model comparison, where the model evidence is approximated using techniques like Variational Bayes or Markov Chain Monte Carlo (MCMC). Connections exhibiting minimal impact on the model evidence – indicated by consistently low posterior probabilities or negligible changes in the free energy – are then pruned from the network. This process directly reduces the number of free parameters requiring estimation, thereby decreasing computational demands and facilitating a more interpretable model by highlighting the most salient neural connections driving observed brain activity. The threshold for pruning is determined empirically, balancing model complexity with goodness-of-fit to the data.

Mapping Disrupted Networks and Predicting Seizure Onset
The Neurological Connectivity and Dynamical Causality Model (NccDCM), leveraging electroencephalography (EEG) data, facilitates the identification of specific alterations within the epileptic network-a crucial step toward understanding how seizures originate and propagate. This computational approach doesn’t merely detect the presence of abnormal activity, but actively maps the disrupted functional connections that characterize the epileptic brain. By pinpointing key nodes and pathways exhibiting atypical behavior, the NccDCM provides a detailed, mechanistic perspective on seizure genesis, moving beyond simple observation to reveal the dynamic interplay of brain regions involved in the epileptic process. This detailed mapping offers the potential to not only refine diagnostic accuracy but also to inform the development of targeted interventions aimed at restoring healthy network function and mitigating seizure activity.
Investigations into the epileptic brain consistently reveal alterations within critical neural networks, notably the Default Mode Network (DMN), thalamus, and caudate nucleus. These structures, typically involved in internal thought, sensory relay, and reward processing respectively, exhibit demonstrably altered functional connectivity patterns in patients experiencing epilepsy. Disruption within the DMN may contribute to the cognitive comorbidities often associated with the condition, while compromised thalamocortical communication can directly impact seizure threshold and propagation. Furthermore, alterations in the caudate nucleus suggest a potential link between epilepsy and changes in motivational or habitual behaviors. These findings underscore the importance of network-level analysis in understanding the complex pathophysiology of epilepsy and point towards potential targets for novel therapeutic interventions designed to restore more typical brain function.
Analysis of dynamic causal nets revealed statistically significant differences between individuals diagnosed with epilepsy and a control group, indicating altered brain network interactions associated with the condition. Furthermore, a clear distinction in network connectivity was observed when comparing brain activity immediately before (preictal) and during (ictal) seizure events – specifically, with a significance level of p < 0.05. These findings suggest that measurable changes in how brain regions influence each other occur both as a chronic feature of epilepsy and as a dynamic process surrounding seizures, offering potential biomarkers for disease characterization and, ultimately, improved predictive capabilities for seizure onset.
Analysis of dynamic causal nets revealed substantial alterations in network connectivity during the transition from the preictal to the ictal phase, with statistically significant differences (p < 0.005) observed across multiple connections. These findings suggest a rapid reorganization of brain networks immediately preceding and during seizures, indicating a breakdown in normal functional integration. The observed shifts aren’t merely a global disruption, but rather involve specific pathways that likely contribute to seizure generation and propagation. This temporal specificity underscores the potential for using these network changes as predictive biomarkers, offering a window into the dynamic processes underlying epileptic events and potentially enabling timely intervention strategies.
The NccDCM’s strength lies in its capacity to address inherent biological variability between individuals, a crucial factor often overlooked in neurological studies. By employing a Mixed-Effects Model, the analysis accounts for unique characteristics of each subject, moving beyond generalized population-level conclusions. This approach minimizes the impact of individual differences on the results, bolstering the reliability and broad applicability of the findings. Consequently, the NccDCM demonstrates improved generalizability, suggesting its potential as a robust tool for identifying biomarkers and ultimately translating research insights into personalized clinical strategies for epilepsy management and prediction.
The pursuit of understanding brain connectivity, as detailed in this novel network of conditionally coupled dynamic causal models, necessitates a willingness to dismantle conventional assumptions. This research doesn’t simply accept established models of EEG data analysis; instead, it actively constructs a new framework, probing the limits of current methodologies. As Ralph Waldo Emerson stated, “Do not go where the path may lead, go instead where there is no path and leave a trail.” The work embodies this sentiment, forging a new path in the analysis of complex neurological data, particularly in identifying disease-related patterns like those seen in epilepsy. The NccDCM approach isn’t about refining existing tools, but rather about reconstructing the very foundations of how we interpret brain activity.
Beyond the Black Box
The presented network of conditionally coupled dynamic causal models-NccDCM-represents a deliberate dismantling of assumptions inherent in traditional brain connectivity analyses. It isn’t simply about finding connections, but about acknowledging where the model itself imposes order on what is, at its core, stochastic. The immediate challenge lies not in scaling the model to larger datasets – though that will inevitably come – but in rigorously testing the limits of its conditional independence assumptions. How much of the identified structure is genuinely reflective of neural dynamics, and how much is an artifact of the model’s architecture? That is the question that must be pursued.
Epilepsy serves as a valuable, albeit unsettling, proving ground. But the true power of NccDCM-and the broader program of reverse-engineering the brain-will be revealed when applied to systems where the ‘ground truth’ is even more elusive. Consider the subtle shifts in connectivity preceding a cognitive decision, or the emergent patterns associated with consciousness itself. These are not pathologies to be diagnosed, but complex systems to be interrogated.
Ultimately, this work isn’t about building a perfect model of the brain-that is a category error. It’s about constructing increasingly sophisticated tools for controlled demolition-carefully dismantling the black box, one assumption at a time, to reveal the beautiful, messy reality within.
Original article: https://arxiv.org/pdf/2601.21478.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Lacari banned on Twitch & Kick after accidentally showing explicit files on notepad
- YouTuber streams himself 24/7 in total isolation for an entire year
- Answer to “A Swiss tradition that bubbles and melts” in Cookie Jam. Let’s solve this riddle!
- Ragnarok X Next Generation Class Tier List (January 2026)
- Adolescence’s Co-Creator Is Making A Lord Of The Flies Show. Everything We Know About The Book-To-Screen Adaptation
- Gold Rate Forecast
- Best Doctor Who Comics (October 2025)
- 2026 Upcoming Games Release Schedule
- Silver Rate Forecast
- 9 TV Shows You Didn’t Know Were Based on Comic Books
2026-01-31 17:49