Author: Denis Avetisyan
New research demonstrates a method for reducing unpredictable variations in clinical risk scores without compromising accuracy or transparency.

This paper introduces a bootstrapping-based regularization technique to improve the stability of individual-level predictions from clinical risk models, including those built with deep learning.
Despite the increasing reliance on deep learning for clinical risk prediction, model instability-where predictions vary substantially with minor data perturbations-remains a critical barrier to adoption. This study introduces a novel regularization framework, detailed in ‘Bootstrapping-based Regularisation for Reducing Individual Prediction Instability in Clinical Risk Prediction Models’, that directly embeds bootstrapping into the training process to constrain prediction variability. By penalizing prediction fluctuations across resampled datasets, the proposed method achieves improved robustness and reproducibility without sacrificing predictive accuracy or interpretability-demonstrated across simulated and clinical datasets including GUSTO-I, Framingham, and SUPPORT. Could this approach offer a practical pathway toward more reliable and trustworthy deep learning models, particularly in data-limited healthcare settings?
The Fragility of Predictive Models: A Matter of Mathematical Purity
Clinical prediction models have become indispensable in modern healthcare, routinely employed to assess patient risk and guide treatment decisions. However, a growing body of evidence reveals a disconcerting fragility: these models frequently demonstrate unpredictable behavior when applied to datasets different from those used in their development. This phenomenon, termed prediction model instability, isn’t merely a matter of slight performance variation; it can manifest as drastically altered risk scores and, consequently, inappropriate clinical recommendations. The issue arises because models learn patterns specific to the training data, failing to generalize effectively to new populations or clinical settings where subtle, yet critical, differences exist. Consequently, a model validated in one hospital system may perform poorly – or even dangerously – when deployed in another, highlighting a crucial need for improved validation techniques and more robust modeling approaches.
The core of many clinical prediction models relies on Empirical Risk Minimisation, a process where algorithms learn to make predictions by identifying patterns within a training dataset. However, this approach inherently involves approximating a complex reality with an incomplete representation of it. The model essentially constructs a simplified version of the world based solely on the data provided, meaning it can struggle when encountering situations not fully captured in that original dataset. This limitation isn’t a flaw in execution, but a fundamental consequence of the methodology; the model extrapolates from observed instances, and any deviation from those instances introduces uncertainty. Consequently, even seemingly minor differences between the training data and the data encountered in a real-world clinical setting can lead to substantial shifts in predictive performance, highlighting the fragility inherent in this widely-used modelling paradigm.
The practical consequences of inconsistent clinical prediction models extend beyond mere statistical concern, directly impacting patient care and fostering a justifiable erosion of trust in these tools. When a model performs well in one setting but falters in another, clinicians understandably become hesitant to rely on its outputs for crucial decisions regarding diagnosis, treatment, and prognosis. This unreliability hinders effective patient risk stratification – the ability to accurately identify individuals most likely to benefit from preventative interventions or require intensive care – potentially leading to delayed or inappropriate treatment. Consequently, there is a growing need for more robust methodologies in model development, focusing on techniques that enhance generalizability, account for data heterogeneity, and provide transparent assessments of predictive uncertainty, ultimately bolstering both clinical confidence and patient outcomes.
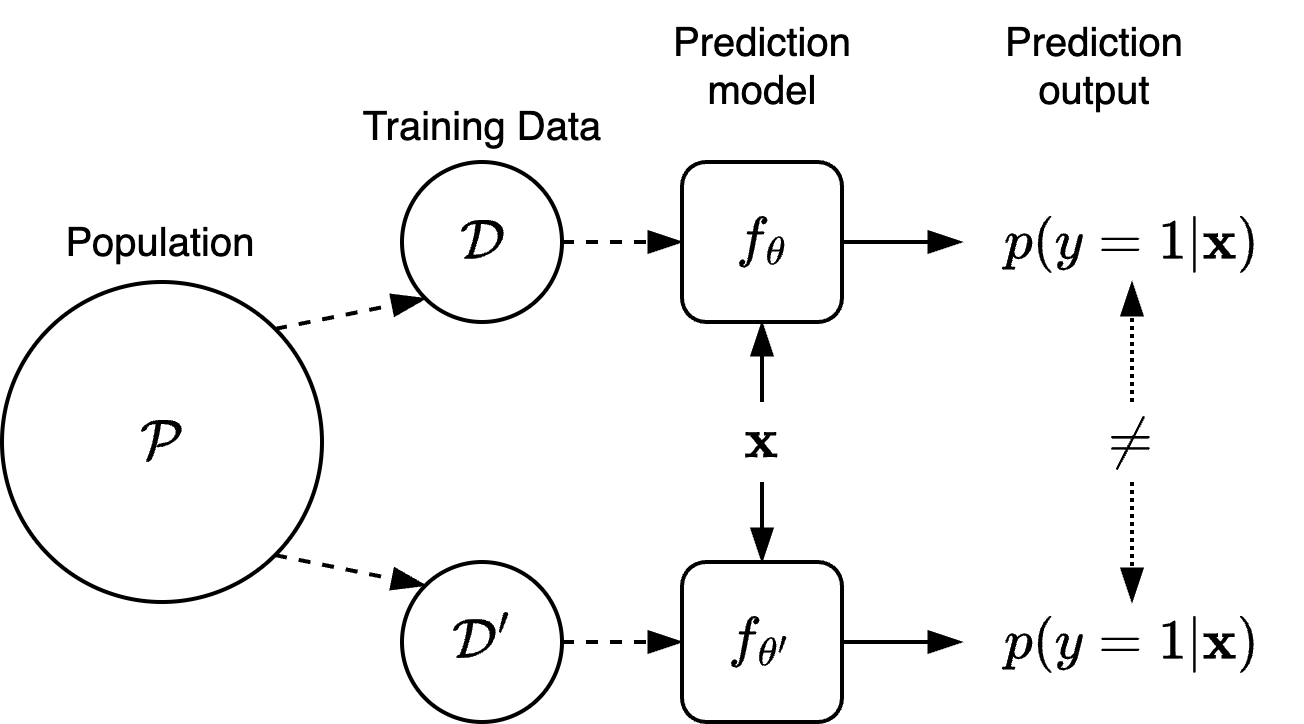
Stability-Based Regularisation: A Principled Approach to Robustness
Stability-based Regularisation integrates the bootstrapping process directly into standard model training. Unlike traditional regularization techniques applied post-training or as separate data augmentation steps, this method repeatedly resamples the training dataset – creating multiple “bootstrap” datasets – and trains the model on each of these resampled sets. The gradients from these multiple training iterations are then aggregated, effectively penalizing model parameters that yield inconsistent predictions across different data subsets. This embedded bootstrapping approach aims to improve model robustness by explicitly encouraging stable and consistent performance rather than simply minimizing error on a single training set.
Bootstrapping, in the context of stability-based regularisation, involves the repeated random sampling of the training dataset with replacement. This creates multiple, slightly different subsets of the original data. A separate model is then trained on each bootstrapped dataset. The principle is that a robust model should yield consistent predictions across these varying subsets; therefore, the training process explicitly encourages the model to minimize the variance of its predictions when exposed to different resampled versions of the training data. This process directly promotes generalization by reducing sensitivity to specific data points and increasing confidence in predictions made on unseen data.
Stability-based regularization demonstrates heightened efficacy when implemented with Deep Learning architectures due to their inherent complexity and large parameter spaces. Deep Neural Networks are prone to overfitting, particularly with limited datasets, leading to poor generalization performance on unseen data. By integrating bootstrapping into the training process, this method mitigates overfitting by enforcing consistency across multiple resampled datasets, effectively smoothing the loss landscape and reducing sensitivity to individual data points. This results in models exhibiting improved resilience to noisy inputs and a greater capacity to generalize to new, previously unobserved examples, ultimately enhancing overall predictive accuracy and reliability.
Stability-based regularisation optimises model parameters by combining Maximum Likelihood Estimation (MLE) with Binary Cross-Entropy Loss. MLE is used to find the parameters that maximise the likelihood of observing the training data, while Binary Cross-Entropy Loss quantifies the difference between predicted probabilities and actual labels. This combined approach doesn’t directly minimise prediction error, but instead aims to reduce the variance in predictions across bootstrap samples. Specifically, the loss function encourages model parameters that yield similar predictions on slightly different resampled datasets, effectively promoting stability and improving generalisation performance. The Binary Cross-Entropy component handles the classification task, while the MLE framework ensures the model consistently assigns high probabilities to the correct classes across these resampled datasets, leading to more robust predictions.
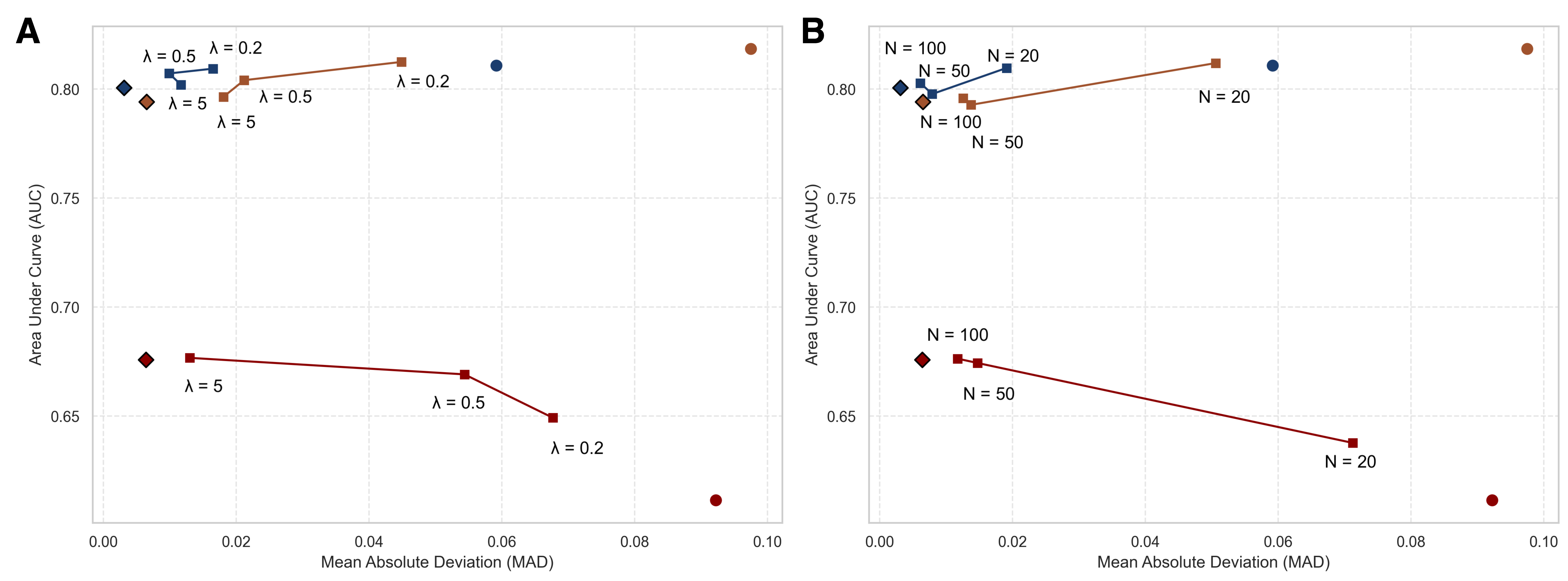
Empirical Validation: Demonstrating Enhanced Stability and Interpretability
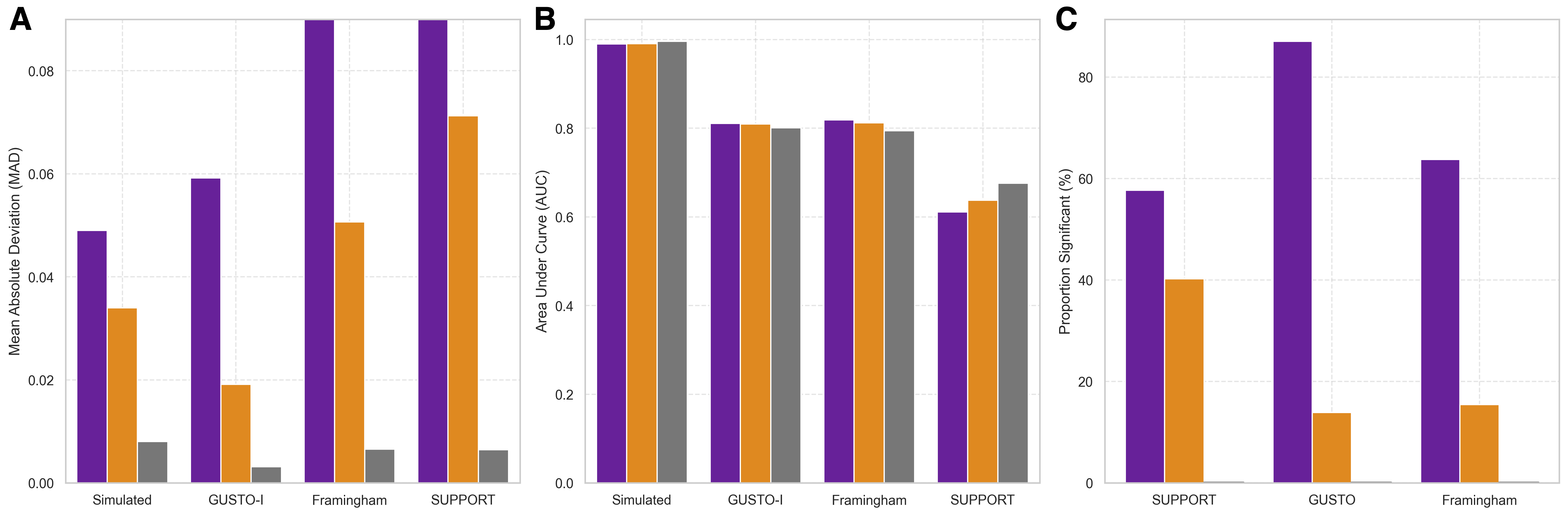
Stability-based Regularisation has been evaluated across multiple datasets – GUSTO-I, Framingham, and SUPPORT – to demonstrate consistent improvements in model robustness. Quantitative results indicate a reduction in Mean Absolute Difference (MAD) when utilizing this regularisation technique. Specifically, the stable model achieved a MAD of 0.034 on a simulated dataset, compared to 0.048 for the standard model. Performance gains were also observed on the GUSTO-I dataset (0.019 vs 0.059), the Framingham dataset (0.057 vs 0.088), and the SUPPORT dataset (0.0712 vs 0.0923), indicating a consistent trend of enhanced predictive stability across diverse datasets.
Model performance was evaluated using both Area Under the Receiver Operating Characteristic Curve (AUC-ROC) and Mean Absolute Difference (MAD) to quantify predictive accuracy and stability. AUC-ROC assesses the model’s ability to discriminate between classes, while MAD provides a measure of the average magnitude of errors in continuous predictions. Comparative analysis across datasets – GUSTO-I, Framingham, and SUPPORT – demonstrated consistent improvements with Stability-based Regularisation; specifically, MAD values were reduced from 0.048 to 0.034 in a simulated dataset, 0.059 to 0.019 on GUSTO-I, 0.088 to 0.057 on Framingham, and 0.0923 to 0.0712 on the SUPPORT dataset, indicating enhanced discriminatory power and reduced prediction error.
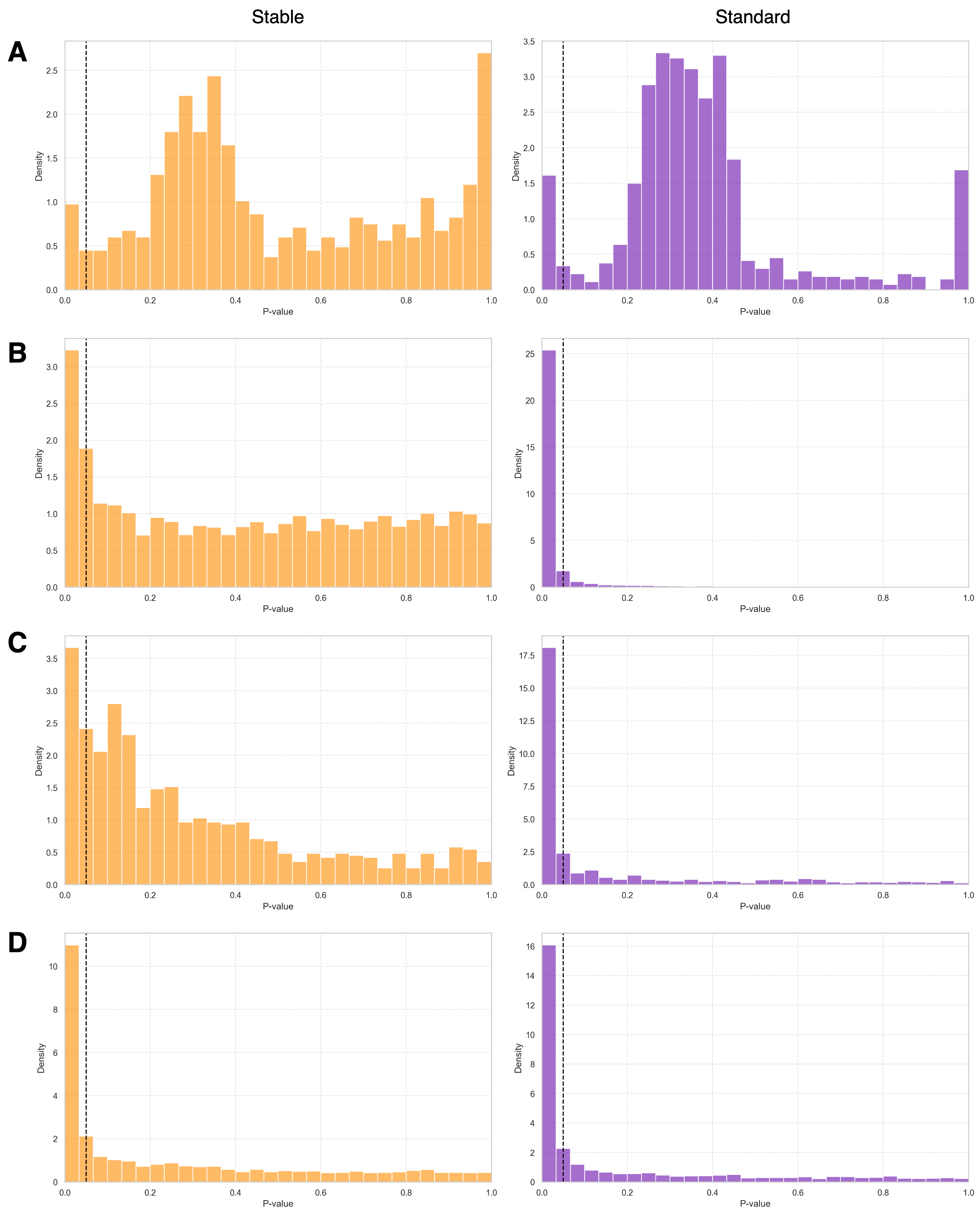
Stability-based Regularisation facilitates model interpretability by enabling the application of techniques such as Shapley Values to determine feature importance and understand the basis for specific predictions. Shapley Values, derived from cooperative game theory, quantify each feature’s contribution to the model’s output, providing a consistent and theoretically grounded explanation for individual predictions. This contrasts with many machine learning models that function as ‘black boxes’, where the reasoning behind predictions remains opaque. By promoting stable and consistent model behavior, this technique allows for more reliable and meaningful application of interpretability methods, offering insights into the model’s decision-making process.
Applying Stability-based Regularisation to ensemble methods, such as Bagging, yields increased predictive consistency. This is achieved by regularising not only the base learners within the ensemble, but also promoting agreement between them. The resultant ensemble demonstrates reduced variance in predictions across different resamples of the training data, leading to more reliable and stable outputs. While specific quantitative comparisons to standard Bagging implementations are not detailed here, the methodology extends the benefits of stability regularization to scenarios where multiple models are combined for prediction.
Quantitative evaluation demonstrates that Stability-based Regularisation consistently reduces prediction error as measured by Mean Absolute Difference (MAD). In a simulated dataset, the stable model achieved a MAD of 0.034, a 14% improvement over the standard model’s MAD of 0.048. Performance gains were also observed across multiple clinical datasets: the stable model attained an MAD of 0.019 on the GUSTO-I dataset, compared to 0.059 for the standard model; on the Framingham dataset, MAD was reduced from 0.088 to 0.057; and on the SUPPORT dataset, the stable model achieved an MAD of 0.0712, improving upon the standard model’s 0.0923.
Towards Reliable Clinical Prediction: A Paradigm Shift in Trust and Accuracy
Personalized risk assessments stand to gain significantly from a new approach that directly addresses individual-level prediction instability. Traditionally, clinical prediction models can yield dramatically different risk scores for the same patient based on slight changes in the data used to train the model-a phenomenon that erodes trust in their accuracy and reliability. This work introduces methods to minimize these fluctuations, ensuring that predictions remain consistent even with minor data variations. By stabilizing these individualized forecasts, clinicians can place greater confidence in identifying patients who would most benefit from preventative measures or tailored treatments, ultimately leading to more informed decision-making and improved patient outcomes. This enhanced stability doesn’t merely refine the accuracy of predictions, but also bolsters the trustworthiness of the entire predictive process.
A significant challenge in clinical prediction lies in the instability of models – their tendency to produce differing results when trained on slightly different datasets. Recent advancements address this by creating models demonstrably less sensitive to minor variations in training data. This robustness translates directly into more consistent predictions, not just within a specific population, but crucially, across diverse patient groups. By minimizing the impact of dataset quirks, these models offer a more generalized and reliable assessment of risk, reducing the potential for inaccurate predictions based on the specific composition of the training cohort. Consequently, healthcare professionals can have increased confidence in the stability and fairness of these predictions, ultimately fostering more equitable and effective patient care.
The development of stability-enhancing techniques in predictive modeling offers a pathway to significantly improve the performance of established clinical risk scores. Current tools like QRisk and EuroSCORE, while widely used, can exhibit inconsistencies due to the inherent variability in patient data and model training. By integrating these new methods, existing Clinical Prediction Models can become more robust and reliable, yielding more accurate assessments of individual patient risk. This refinement extends beyond cardiology and general practice-the principles apply across numerous medical specialties, from oncology and surgery to critical care-potentially leading to more informed treatment decisions, optimized resource allocation, and, ultimately, improved patient outcomes through proactive and personalized care strategies.
The convergence of Stability-based Regularisation and Shapley Values offers a powerful mechanism for dissecting the factors driving clinical predictions. This approach not only enhances the reliability of models but also illuminates which variables are most influential in determining risk. Stability-based Regularisation ensures that small changes in the training data don’t drastically alter the model’s core logic, while Shapley Values, originating from game theory, fairly distribute credit among these variables based on their contribution to the prediction. Consequently, clinicians gain a clearer understanding of the key predictive factors – be it age, blood pressure, or genetic markers – allowing for more precise diagnoses and, crucially, the development of targeted interventions designed to address the most impactful elements of a patient’s risk profile. This improved interpretability moves beyond simply forecasting outcomes to informing proactive and personalized healthcare strategies.
The pursuit of robust and reliable prediction models, as detailed in the article, aligns with a fundamental principle of computational correctness. Ken Thompson famously stated, “Debugging is twice as hard as writing the code in the first place. Therefore, if you write the code as cleverly as possible, you are, by definition, not smart enough to debug it.” This sentiment underscores the importance of simplicity and provability in system design. The bootstrapping-based regularization technique presented seeks to minimize individual prediction instability-essentially, ensuring a consistent and reproducible outcome. Just as Thompson advocates for avoiding overly complex code, this method champions a model that prioritizes stability without sacrificing accuracy, aiming for a solution that is demonstrably correct, not merely appearing to work on limited tests.
What Remains Invariant?
The pursuit of stable prediction models in clinical settings, as demonstrated by this work, inevitably circles back to a fundamental question: Let N approach infinity – what remains invariant? While bootstrapping offers a practical method for reducing individual prediction instability, it addresses a symptom, not the underlying disease. The true instability stems from the inherent limitations of finite datasets and the inductive biases embedded within any model, however ‘deep’ or ‘regularized’. The presented technique, admirable in its preservation of interpretability, merely smooths the variance; it does not fundamentally alter the model’s sensitivity to unseen, yet plausible, data points.
Future work must move beyond variance reduction and grapple with the question of model robustness. Can techniques from formal verification – proofs of correctness – be adapted to clinical prediction? Perhaps a more fruitful avenue lies in explicitly modelling uncertainty – not as a nuisance parameter to be minimized, but as a first-class citizen of the predictive process. SHAP values, while useful for post-hoc explanation, remain descriptive; a truly elegant solution would incorporate uncertainty directly into the model’s architecture.
The current emphasis on predictive accuracy, divorced from considerations of epistemic risk, is a precarious path. A model that performs admirably on held-out data but collapses spectacularly when faced with a novel outlier is, ultimately, a failure of mathematical principle. The challenge, then, is not simply to predict better, but to understand when and why a prediction can be trusted – a question that demands a return to first principles.
Original article: https://arxiv.org/pdf/2602.11360.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Movie Games responds to DDS creator’s claims with $1.2M fine, saying they aren’t valid
- SHIB PREDICTION. SHIB cryptocurrency
- Scream 7 Will Officially Bring Back 5 Major Actors from the First Movie
- United Airlines can now kick passengers off flights and ban them for not using headphones
- The MCU’s Mandarin Twist, Explained
- These are the 25 best PlayStation 5 games
- Rob Reiner’s Son Officially Charged With First Degree Murder
- Server and login issues in Escape from Tarkov (EfT). Error 213, 418 or “there is no game with name eft” are common. Developers are working on the fix
- MNT PREDICTION. MNT cryptocurrency
- Gold Rate Forecast
2026-02-14 13:13