Author: Denis Avetisyan
A new self-supervised learning approach is dramatically improving the analysis of X-ray angiograms, enabling more accurate vascular segmentation and detection.
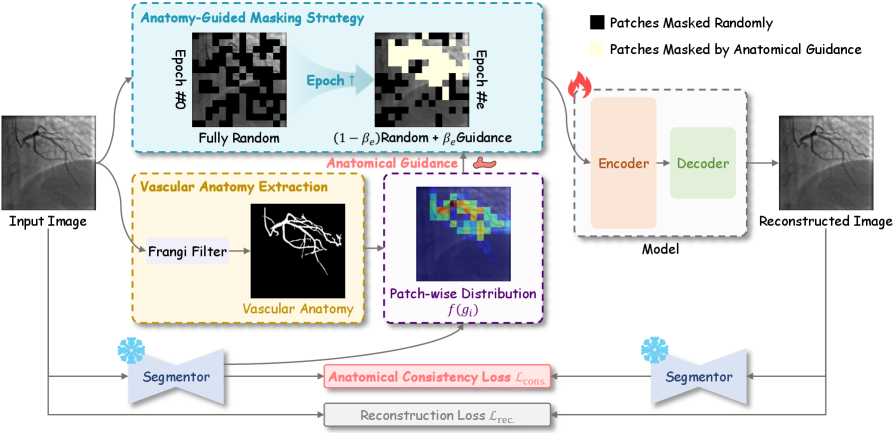
Researchers introduce VasoMIM, a framework leveraging anatomical guidance and masked image modeling for state-of-the-art performance in X-ray angiogram analysis.
Despite the clinical importance of X-ray angiography for diagnosing cardiovascular diseases, current deep learning approaches are hampered by limited annotated data. This work introduces a novel framework for ‘Vascular anatomy-aware self-supervised pre-training for X-ray angiogram analysis’-VasoMIM-which leverages anatomical knowledge and a large-scale dataset, XA-170K, to significantly improve performance on downstream tasks. By strategically masking vessel-containing patches and enforcing anatomical consistency, VasoMIM learns robust vascular representations, achieving state-of-the-art results in vascular segmentation and detection. Could this anatomy-aware pre-training strategy serve as a foundational step towards fully automated and accurate cardiovascular disease diagnosis?
The Illusion of Clarity: Visualizing Vascular Networks
The persistent prevalence of cardiovascular diseases as a primary cause of death globally underscores an urgent need for advancements in diagnostic capabilities. These conditions, encompassing a range of ailments from coronary artery disease to stroke, frequently necessitate precise anatomical understanding for effective intervention. Despite progress in medical technology, timely and accurate diagnosis remains a significant challenge, often hampered by the limitations of current imaging techniques and the subtle nature of early-stage vascular dysfunction. Consequently, research is heavily focused on developing tools that can not only detect these diseases earlier but also provide clinicians with detailed visualizations of the circulatory system, ultimately improving patient outcomes and reducing the substantial burden these conditions place on healthcare systems worldwide.
Precise depiction of vascular anatomy through X-ray Angiography forms the cornerstone of effective cardiovascular diagnosis and intervention. This imaging technique allows clinicians to non-invasively assess blood vessel structure, identifying critical blockages, aneurysms, or malformations that might otherwise remain undetected. The ability to visualize these intricate networks is paramount for planning complex procedures like angioplasty, stenting, or bypass surgery, enabling surgeons to navigate with greater accuracy and minimize patient risk. Beyond treatment, detailed angiographic visualization aids in early disease detection, allowing for preventative measures and improved patient outcomes; it provides a roadmap for understanding and addressing the complexities of the circulatory system, ultimately impacting the quality of care delivered to individuals with cardiovascular conditions.
Conventional techniques for delineating blood vessels in medical imagery often falter when faced with the inherent intricacies of vascular anatomy. The human circulatory system presents a formidable challenge to automated segmentation algorithms due to the sheer density of vessels, their varying sizes and opacities, and the presence of anatomical variations unique to each individual. Existing methods, frequently reliant on identifying edges or tracking intensity gradients, struggle to differentiate between actual vessel walls and surrounding tissue noise, leading to both false positives and missed detections. Furthermore, subtle differences in contrast, caused by patient-specific factors or imaging limitations, can obscure vessel boundaries, while the tortuous and branching nature of capillaries and smaller arterioles further complicates accurate reconstruction of the complete vascular network. This limitation necessitates the development of more robust and adaptable image processing techniques capable of overcoming these challenges and providing clinicians with precise, high-resolution visualizations of the vasculature.
Self-Supervision: A Necessary Compromise
Self-Supervised Learning (SSL) provides a methodology for training deep learning models on unlabeled datasets, circumventing the need for extensive manual annotation. Traditional supervised learning requires paired input data with corresponding labels, which is often a limiting factor, particularly in medical imaging. SSL techniques instead formulate pretext tasks – artificial prediction problems derived from the data itself – allowing the model to learn meaningful representations by solving these tasks. The learned representations capture inherent data structure and can then be transferred to downstream tasks with limited labeled data, offering a pathway to improved performance and reduced annotation burden. This is achieved by leveraging the inherent redundancy and relationships within the data to create supervisory signals, effectively allowing the model to “learn from itself.”
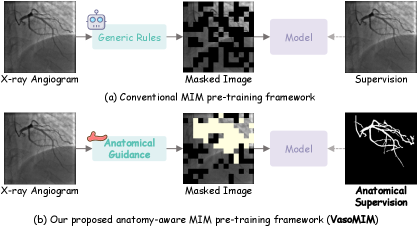
Masked Image Modeling (MIM) operates on the principle of artificially masking portions of an input image and training a model to reconstruct the missing pixel values. This process forces the model to learn contextual relationships within the data to accurately predict the masked regions. Specifically, an input image is partially obscured, and the model attempts to fill in these gaps based on the visible portions, effectively learning a representation of the underlying data distribution. The reconstruction task is typically performed using an encoder-decoder architecture, where the encoder compresses the visible image into a latent representation, and the decoder reconstructs the complete image, including the masked areas. The efficacy of MIM relies on the model’s ability to capture dependencies between different image regions, thereby learning robust and generalizable feature representations without requiring explicit labels.
Direct application of Masked Image Modeling (MIM) to medical images frequently yields suboptimal performance due to characteristics unique to this domain. Medical images often exhibit low contrast, high noise, and substantial anatomical variations between patients. Furthermore, the relatively small receptive field of standard MIM architectures struggles to capture long-range dependencies critical for understanding complex anatomical structures. Unlike natural images with consistent semantic content, medical images contain subtle, high-frequency details essential for accurate diagnosis, which are easily lost during masking and reconstruction. These factors combine to make learning robust representations with standard MIM more challenging in medical imaging than in natural image domains.
VasoMIM is a self-supervised learning framework designed to improve pre-training for medical imaging by leveraging known vascular anatomy. The methodology incorporates anatomical priors – specifically, segmentation maps indicating the location of blood vessels – into the masking and reconstruction process of Masked Image Modeling. Rather than randomly masking image patches, VasoMIM strategically masks regions informed by vascular structure, forcing the model to learn representations that are sensitive to anatomical relationships. This is achieved by weighting the reconstruction loss based on the overlap between masked regions and vascular structures, effectively prioritizing the accurate reconstruction of vessel anatomy during pre-training. The resulting model demonstrates improved performance on downstream tasks, particularly those involving vascular segmentation and analysis, compared to standard MIM approaches applied directly to medical images.
Anatomical Fidelity: A Fragile Illusion of Control
The Anatomical Consistency Loss implemented within VasoMIM functions by penalizing deviations from expected vascular topology during reconstruction. This loss component operates on the principle that vascular structures adhere to specific anatomical constraints; therefore, the model is trained to prioritize outputs that exhibit these characteristics. Specifically, the loss calculates discrepancies between the predicted vascular segmentation and an established model of plausible vascular anatomy, encouraging the network to generate reconstructions that are topologically sound and biologically realistic. This approach is critical for mitigating artifacts and ensuring the clinical validity of the reconstructed vasculature, even in cases of incomplete or noisy input data.
The Anatomical Consistency Loss employed within VasoMIM utilizes the UNeXt-S architecture to enforce realistic topological constraints during vascular segmentation. This loss function operates by penalizing reconstructions that deviate from expected anatomical configurations, effectively promoting the generation of vascular structures that align with established biological principles. Specifically, it encourages connectivity and discourages implausible branching or discontinuities, leading to improved segmentation accuracy and the production of anatomically consistent vascular models. The UNeXt-S foundation provides a robust feature extraction capability essential for accurately assessing and enforcing these anatomical plausibility criteria.
The VasoMIM framework utilizes the XA-170170K dataset for pre-training, a collection of 170,170 X-ray angiogram images. This dataset is currently the largest publicly available resource of its kind, providing a substantial foundation for the model’s vascular segmentation capabilities. The scale of XA-170170K is critical, enabling the framework to learn robust features and generalize effectively to new angiographic data. Data from this source significantly contributes to the anatomical consistency and performance improvements observed in the reconstructed vascular structures.
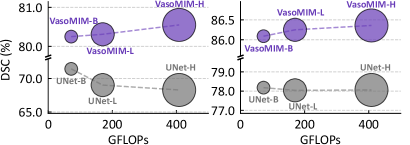
Performance gains were achieved through a combination of data scaling and model scaling techniques. Utilizing these methods, the VasoMIM framework attained a Dice Similarity Coefficient (DSC) of 76.01% when trained with only 10% of the available labeled data. This result represents a 0.50% improvement over the performance of TransUNet, a leading fully supervised baseline, which required training on the complete 100% of the labeled dataset to achieve a DSC of 75.51%. These findings demonstrate the effectiveness of the scaling strategies in maximizing performance with limited labeled data.
Beyond the Map: Towards Pragmatic Stenosis Detection
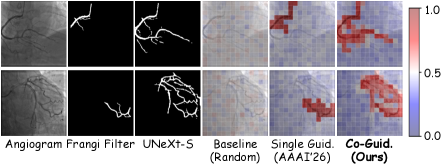
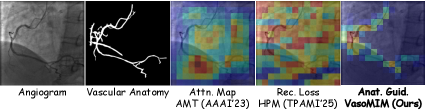
VasoMIM leverages the Frangi Filter, a sophisticated technique rooted in the analysis of the Hessian Matrix, to meticulously delineate vascular anatomy. The Hessian Matrix, a matrix of second-order partial derivatives, allows the filter to identify and enhance vessel-like structures by assessing local image curvature and responsiveness to different orientations. This process effectively highlights ridge-like features characteristic of blood vessels while suppressing noise and irrelevant details. By accurately mapping the vessel network, VasoMIM establishes a crucial foundation for subsequent analysis, enabling precise identification of subtle changes in vessel diameter and the detection of potential stenoses – narrowings that can impede blood flow. The filter’s ability to robustly extract these anatomical features is central to VasoMIM’s performance and contributes significantly to its diagnostic capabilities.
A precise understanding of vascular anatomy is foundational to the reliable detection of stenosis – the narrowing of blood vessels. Accurate stenosis detection isn’t simply about identifying a constriction; it requires contextual awareness of the vessel’s typical diameter, branching patterns, and overall morphology. Without this anatomical baseline, differentiating between normal physiological variations and pathological narrowing becomes exceedingly difficult, leading to false positives or missed diagnoses. Consequently, methods that prioritize robust vessel segmentation and anatomical reconstruction, like VasoMIM, provide a crucial advantage, enabling more confident and precise identification of stenotic lesions and ultimately improving the efficacy of cardiovascular disease diagnosis and treatment planning.
Evaluations demonstrate that VasoMIM significantly enhances the precision of stenosis detection, achieving gains of +1.32% on the mAP50 metric, +0.96% on mAP75, and an overall improvement of +0.83% compared to existing self-supervised learning (SSL) baselines. These metrics, commonly used to assess the accuracy of object detection algorithms, indicate a substantial increase in the ability to correctly identify and localize narrowings within blood vessels. The observed improvements suggest that VasoMIM’s anatomical understanding, derived from its vascular anatomy extraction process, provides a more robust foundation for pinpointing stenoses, ultimately leading to more reliable diagnostic outcomes.
VasoMIM establishes a robust groundwork for pinpointing vascular anomalies by first creating a detailed map of vessel structure. Accurate vessel segmentation – the process of precisely outlining blood vessels – is paramount for detecting subtle changes indicative of stenosis, or narrowing. The system’s capacity to delineate these vessels with high fidelity allows for the subsequent identification of constrictions and blockages that might otherwise be missed. This detailed anatomical understanding doesn’t simply highlight where vessels are, but also enables the quantification of changes in vessel diameter, crucial for assessing the severity of stenosis and informing clinical decision-making. Ultimately, this enhanced capacity to detect narrowings promises earlier and more accurate diagnoses of cardiovascular disease, potentially leading to more effective interventions and improved patient outcomes.
The potential for more accurate stenosis detection extends directly to improved cardiovascular care. Precise identification of narrowed vessels – often indicative of atherosclerosis or other vascular diseases – allows clinicians to move beyond generalized diagnoses and formulate targeted treatment plans. Earlier and more definitive detection, facilitated by tools like VasoMIM, promises to reduce the incidence of severe cardiovascular events, such as stroke or heart attack. This proactive approach enables timely interventions – from lifestyle modifications and medication to minimally invasive procedures – ultimately enhancing patient outcomes and reducing the long-term burden of cardiovascular disease. The capability to pinpoint the location and severity of stenoses with greater confidence represents a significant step forward in preventative cardiology and vascular health management.
The pursuit of elegant solutions in medical imaging invariably meets the harsh reality of clinical data. This work, with its VasoMIM framework and anatomical guidance, attempts to impose order on the chaos of X-ray angiograms, a noble effort, to be sure. Yet, one suspects even meticulously crafted pre-training, leveraging self-supervised learning and masked image modeling, will eventually succumb to the unpredictable variations found in production. As Yann LeCun once noted, “If we want to be serious about artificial intelligence, we need to get better at building systems that can learn from very little data.” The promise of foundation models is enticing, but the long-term viability hinges on robustness, and robustness, inevitably, is tested by the unexpected. One anticipates a future filled with increasingly sophisticated architectures, each destined to become tomorrow’s technical debt.
What’s Next?
The enthusiasm for ‘foundation models’ in medical imaging will inevitably collide with the realities of clinical practice. This work, while demonstrating gains in vascular segmentation – a problem that hasn’t fundamentally changed in decades – merely shifts the complexity. The anatomical guidance is clever, certainly, but it feels less like intelligence and more like hard-coding what a reasonably trained radiology technician already knows. The real challenge isn’t mimicking segmentation; it’s handling the ambiguous cases, the anomalies, the patient-specific quirks that no dataset, however large, can fully capture. If a system crashes consistently, at least it’s predictable.
Future iterations will likely focus on integrating this pre-training with downstream tasks, chasing ever-smaller incremental improvements. A more fruitful, though less glamorous, avenue might be focusing on data efficiency. The scale of these pre-training datasets is becoming unsustainable. Perhaps the value isn’t in bigger data, but in better data – carefully curated, actively maintained, and acknowledging that labelled data is, ultimately, a human expense. The ‘cloud-native’ architecture feels largely cosmetic; the same mess, just more expensive.
Ultimately, this research, like most, creates as many questions as it answers. The focus on automated segmentation feels strangely detached from the actual clinical workflow. The unspoken truth is, these models aren’t replacing radiologists; they’re generating notes for digital archaeologists who will one day try to decipher why this particular algorithm failed on patient X.
Original article: https://arxiv.org/pdf/2602.11536.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- United Airlines can now kick passengers off flights and ban them for not using headphones
- SHIB PREDICTION. SHIB cryptocurrency
- Movie Games responds to DDS creator’s claims with $1.2M fine, saying they aren’t valid
- Scream 7 Will Officially Bring Back 5 Major Actors from the First Movie
- These are the 25 best PlayStation 5 games
- The MCU’s Mandarin Twist, Explained
- Rob Reiner’s Son Officially Charged With First Degree Murder
- Pacific Drive’s Delorean Mod: A Time-Traveling Adventure Awaits!
- MNT PREDICTION. MNT cryptocurrency
- Gold Rate Forecast
2026-02-15 23:05