Author: Denis Avetisyan
Researchers are leveraging the unique folding patterns of the brain to build more accurate diagnostic tools for Alzheimer’s and Lewy body dementia.
A probability-invariant random walk learning framework on gyral folding-based cortical similarity networks improves classification of neurodegenerative diseases.
Differentiating Alzheimer’s disease from Lewy body dementia remains challenging due to overlapping clinical presentations, yet accurate diagnosis is crucial for targeted intervention. This work, ‘Probability-Invariant Random Walk Learning on Gyral Folding-Based Cortical Similarity Networks for Alzheimer’s and Lewy Body Dementia Diagnosis’, introduces a novel framework leveraging individualized cortical folding patterns to improve dementia classification. By representing brain networks as distributions of anonymized random walks-and ensuring permutation invariance-the proposed method overcomes limitations of traditional graph learning approaches sensitive to anatomical variability. Could this anatomy-aware approach offer a more robust and personalized pathway toward earlier and more accurate dementia diagnosis?
Mapping Cortical Architecture: A Foundation for Network Integrity
Historically, brain imaging techniques have frequently prioritized identifying large anatomical regions, often overlooking the nuanced intricacies of cortical structure. This approach, while useful for broad localization, can obscure vital individual differences in brain organization and function, as the precise patterns of folding-the gyri and sulci-vary considerably between individuals. These subtle variations aren’t merely anatomical quirks; they represent differences in cortical surface area, neuronal connectivity, and ultimately, cognitive processing efficiency. By concentrating on broad regions, researchers risk losing sight of these critical details, hindering a complete understanding of how brain structure relates to behavior and potentially obscuring the biological basis of neurological disorders. A shift towards methods that capture these fine-grained structural characteristics is therefore essential for advancing the field of neuroanatomy and cognitive neuroscience.
The human brain’s remarkable complexity isn’t simply a matter of size, but rather the intricate folding of its outer layer, the cortex. These folds – gyri and sulci – aren’t random; they define a unique network crucial for individual cognitive abilities and neurological health. Accurate mapping of this gyral network allows researchers to move beyond broad regional analyses and pinpoint subtle variations in cortical structure that correlate with differences in intelligence, personality, and susceptibility to brain disorders. This detailed cartography reveals how efficiently information flows within the brain, and how disruptions to this network can manifest as cognitive deficits or neurological symptoms. By precisely characterizing these individual patterns of cortical folding, scientists gain a more nuanced understanding of the brain’s organization and its impact on behavior.
Detailed mapping of the brain’s intricate architecture now benefits from sophisticated software like FreeSurfer, which enables researchers to computationally reconstruct the cortical surface with unprecedented precision. This tool doesn’t simply identify broad brain regions; instead, it meticulously traces the complex folds – gyri and sulci – creating a digital model of the individual’s unique cortical network. The resulting reconstruction provides a far richer substrate for analysis than traditional methods, allowing for the investigation of subtle structural variations linked to cognitive abilities, neurological disorders, and individual differences. By quantifying the geometry of these cortical networks-including surface area, volume, and the complexity of folding-FreeSurfer facilitates a deeper understanding of how brain structure relates to function and behavior, opening new avenues for both basic neuroscience and clinical applications.
PaIRWaL: A Probabilistic Framework for Gyral Network Classification
PaIRWaL utilizes a Probability-Invariant Random-Walk Learning (PIRWL) approach for the classification of gyral folding networks, differing from traditional graph learning methods by focusing on the probabilistic properties of random walks rather than node or edge features. This is achieved by representing random walks as sequences of events, allowing the framework to capture structural information inherent in network topology. The probability invariance is critical as it mitigates the impact of isomorphic variations within gyral networks, which often present similar functional characteristics despite differing structural arrangements. This approach facilitates a more robust and sensitive classification process, particularly relevant for identifying subtle differences in brain network organization that may be indicative of neurological conditions.
The PaIRWaL framework enhances structural information capture by transforming random walks on gyral folding networks into event sequences that incorporate anatomical priors. These priors are derived from pre-defined Regions of Interest (ROIs), effectively weighting the random walk based on the functional or anatomical significance of specific brain areas. This process moves beyond simple path enumeration by associating each network node transition with ROI membership, creating a richer representation of the walk’s trajectory. Consequently, the framework doesn’t just record where a walk goes, but also through which anatomically defined regions, thereby increasing sensitivity to nuanced structural characteristics and improving classification accuracy.
The PaIRWaL framework employs a Minimum Degree Local Rule during random walk sampling to mitigate bias and ensure comprehensive network exploration. This rule prioritizes nodes with lower degrees during the walk, preventing the process from becoming trapped in high-degree hubs and promoting traversal to less-visited regions. The selection process is further refined by incorporating edge Conductance – a measure of the minimum cut between a node’s neighborhood and the rest of the graph – which helps to identify and prioritize edges that facilitate balanced exploration and prevent walks from prematurely terminating in isolated subgraphs. This approach effectively addresses potential imbalances in random walk sampling that can occur in brain networks with heterogeneous degree distributions.
PaIRWaL directly mitigates the problem of Graph Isomorphism – the challenge of distinguishing between graphs that appear structurally different despite possessing identical topological properties – by incorporating anatomical priors and a probability-invariant learning approach. This design allows the framework to focus on functionally relevant network features rather than being misled by isomorphic representations. Quantitative evaluations demonstrate that PaIRWaL achieves consistently higher classification accuracy across multiple benchmark datasets compared to state-of-the-art graph classification methods, including those based on graph kernels and deep learning, specifically in tasks requiring the differentiation of subtle structural variations in gyral folding networks.
Differentiating Lewy Body Dementia: A Validation of the PaIRWaL Framework
Lewy Body Dementia (LBD) presents diagnostic challenges due to substantial clinical and pathological overlap with Alzheimer’s Disease (AD), frequently leading to misdiagnosis. The PaIRWaL framework offers a potential solution by leveraging subtle variations in gyral cortical folding patterns to differentiate LBD brains from those of cognitively normal controls and those affected by AD. This approach moves beyond traditional volumetric or morphometric analyses, focusing instead on the geometric complexity of cortical surfaces. PaIRWaL’s classification capabilities have demonstrated superior accuracy across multiple evaluation tasks, including a multiclass comparison of AD, LBD, and controls, as well as binary comparisons between each disease state and controls, suggesting its utility as a supplementary diagnostic tool.
The PaIRWaL framework enhances differentiation between Lewy Body Dementia (LBD) and Cognitively Normal Controls (CN) by analyzing subtle variations in gyral folding patterns. Evaluations across multiple classification tasks – including a three-way comparison of Alzheimer’s Disease, LBD, and CN, as well as pairwise comparisons of AD vs CN, LBD vs CN, and AD vs LBD – demonstrate that this approach achieves the highest reported accuracy rates to date. This improved performance is attributed to the framework’s sensitivity to nuanced structural differences not readily captured by conventional methods, allowing for more precise identification of LBD-related cortical alterations.
Analysis of data from the Multimodal Imaging in Lewy Body Disorders project confirms the efficacy of the PaIRWaL framework in identifying structural brain signatures indicative of Lewy Body Dementia. Comparative analysis demonstrated statistically significant improvements (p < 0.05) in differentiating Lewy Body Dementia from control groups and Alzheimer’s Disease when utilizing the PaIRWaL framework as opposed to baseline models. These results indicate that the framework successfully captures subtle, disease-specific variations in brain morphology as evidenced by the multimodal imaging data, providing a quantifiable basis for improved diagnostic accuracy.
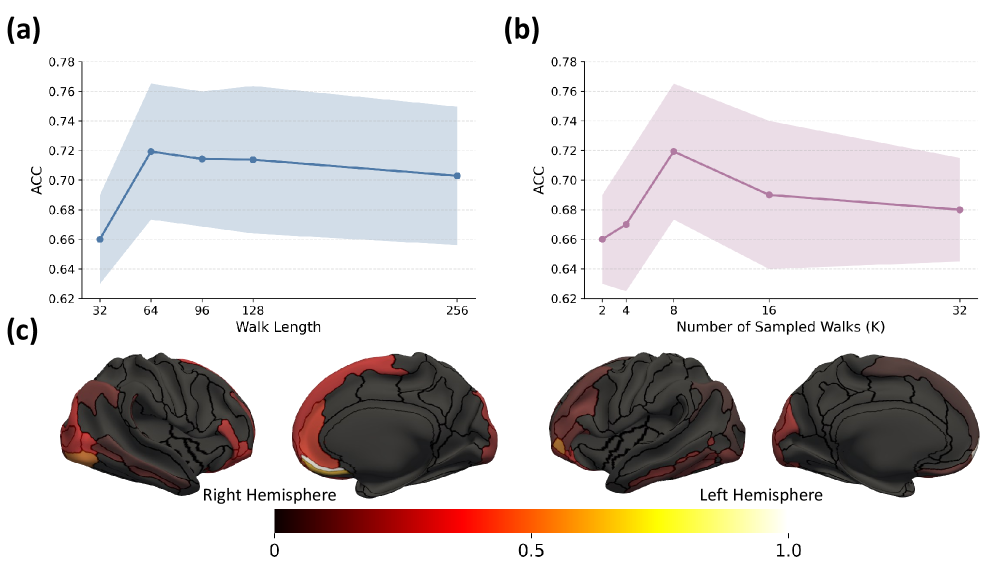
The PaIRWaL framework achieved peak performance in differentiating Lewy Body Dementia when utilizing a random walk length between 64 and 128 steps, coupled with 8 sampled random walks per subject. This parameter configuration facilitated efficient estimation of the underlying cortical graph structures relevant to disease identification. Shorter walk lengths proved insufficient to capture broader structural relationships, while lengths exceeding 128 did not yield significant improvements in classification accuracy. The use of 8 sampled walks balanced computational cost with the robustness of the estimated graph, providing a statistically reliable representation of individual brain morphology for improved diagnostic differentiation.
Expanding the Network Perspective: Towards a Comprehensive Understanding of Brain Organization
Representing the brain as a network, or graph, fundamentally shifts how researchers approach neurological study. This perspective treats brain regions as nodes and the connections between them – whether structural or functional – as edges, enabling the application of powerful network science tools. This analytical framework moves beyond simply identifying where activity occurs, and instead focuses on how different areas interact and contribute to complex cognitive processes. By quantifying network properties like centrality, clustering, and path length, scientists can characterize the brain’s organizational principles and identify disruptions associated with neurological disorders. The graph-based approach allows for a more holistic understanding of brain function, revealing how localized changes can propagate across the entire network and impact overall cognitive performance. Ultimately, this method facilitates the development of more sophisticated models of brain function and offers new avenues for diagnosing and treating neurological conditions.
A powerful synergy emerges when diverse brain network analysis techniques are combined with the PaIRWaL framework, offering a more nuanced understanding of brain organization. Methods like BrainPrint, which captures unique patterns of brain connectivity, and Atlas-Based Brain Networks – leveraging detailed anatomical references like the Destrieux Atlas – provide a structural foundation. Complementary approaches, such as Minimum Spanning Tree analysis – identifying the most critical connections within the network – and Kullback-Leibler Divergence – quantifying the difference between brain network distributions – add layers of functional and comparative insight. Integrating these tools with PaIRWaL not only refines the precision of network characterization, but also unlocks the potential to explore individual brain differences and the intricate relationships between brain structure and cognitive processes with unprecedented detail.
The application of advanced graph neural networks-including GraphSAGE, Graph Attention Networks (GAT), Graph Isomorphism Network (GIN), and BrainNetCNN-significantly enhances the capacity to dissect the intricacies of brain data represented as networks. These algorithms move beyond traditional node-based analysis by considering relationships between brain regions, allowing for a more holistic understanding of functional connectivity. GraphSAGE, for example, efficiently generates node embeddings by sampling and aggregating features from a node’s neighborhood, while GAT leverages attention mechanisms to weigh the importance of different connections. GIN focuses on maximizing the discriminative power of graph representations, and BrainNetCNN is specifically designed to capture spatiotemporal patterns in brain activity. Collectively, these methods empower researchers to identify subtle variations in brain organization, predict cognitive performance, and ultimately, gain deeper insights into the biological basis of behavior by effectively leveraging the inherent relational structure of neural data.
Beyond simply identifying patterns associated with neurological disorders, representing the brain as a complex network allows researchers to explore the unique architecture of each individual’s brain. This shift in perspective moves beyond group averages, revealing the subtle variations that contribute to diverse cognitive abilities and behavioral traits. By analyzing the intricate connections within a person’s brain network, scientists can begin to decipher how specific network properties – such as efficiency, modularity, and resilience – relate to individual differences in learning, memory, and decision-making. This detailed understanding of brain organization promises to unlock the neural basis of cognition, paving the way for personalized approaches to cognitive enhancement and interventions tailored to an individual’s unique brain profile.
The pursuit of robust diagnostic tools, as demonstrated by PaIRWaL, inherently demands a focus on invariant properties. The framework’s reliance on probability-invariant random walks across gyral folding networks exemplifies this principle; it seeks to distill signal from noise by identifying characteristics that remain consistent despite individual brain variation. This resonates with the philosophical insight of Georg Wilhelm Friedrich Hegel: “We must grasp the truth that the real is rational and the rational is real.” The algorithm, in its search for underlying patterns unaffected by superficial differences, attempts to reveal the ‘rational’ structure within the ‘real’ complexity of neurodegenerative disease, striving for a classification that is fundamentally grounded in invariant principles – a provable, rather than merely observed, correlation.
What Remains to Be Proven?
The presented framework, while demonstrating a practical efficacy in classification, sidesteps a fundamental question. The reliance on gyral folding networks, however biologically inspired, remains largely descriptive. A truly elegant solution would derive these networks from first principles – a mathematically rigorous model of cortical development and degradation. The current approach, though achieving robust results, operates within the constraints of observed morphology, failing to predict why certain folding patterns correlate with disease states. This is not prediction, but pattern recognition.
Further exploration must address the limits of probability invariance. While the method mitigates variability, it does not eliminate it. The assumption that random walks, even probability-invariant ones, fully capture the complexity of neural information flow is, at best, a simplification. The deterministic underpinnings of cognitive decline are likely far more nuanced. Reproducibility, a cornerstone of scientific validity, demands a deeper understanding of the method’s sensitivity to parameter selection and data preprocessing – areas currently lacking detailed examination.
The true test lies not in achieving higher accuracy on benchmark datasets, but in developing a framework capable of generalization. Can this approach be extended to predict disease progression, or to differentiate between subtypes of dementia with greater precision? Until the method moves beyond correlation and approaches causation, it remains a sophisticated tool, but not a complete theory. The pursuit of mathematical purity continues.
Original article: https://arxiv.org/pdf/2602.17557.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- All Golden Ball Locations in Yakuza Kiwami 3 & Dark Ties
- NBA 2K26 Season 5 Adds College Themed Content
- All Itzaland Animal Locations in Infinity Nikki
- Elder Scrolls 6 Has to Overcome an RPG Problem That Bethesda Has Made With Recent Games
- What time is the Single’s Inferno Season 5 reunion on Netflix?
- Hollywood is using “bounty hunters” to track AI companies misusing IP
- Gold Rate Forecast
- Exclusive: First Look At PAW Patrol: The Dino Movie Toys
- Star Trek’s Controversial Spock Romance Fixes 2 Classic TOS Episodes Fans Thought It Broke
- Heated Rivalry Adapts the Book’s Sex Scenes Beat by Beat
2026-02-22 23:24