Author: Denis Avetisyan
Researchers have developed a novel machine learning method for identifying sepsis earlier and with greater clarity by leveraging the power of relational data.
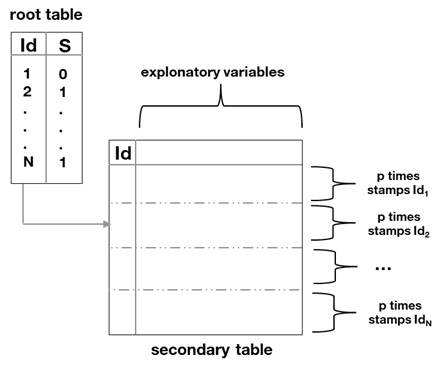
This review details a fully interpretable model utilizing Bayesian classification, Minimum Description Length feature selection, and time-series analysis of relational sepsis data.
Despite advances in critical care, early and interpretable prediction of sepsis remains a significant clinical challenge due to its complex and heterogeneous nature. This is addressed in ‘Temporal Sepsis Modeling: a Fully Interpretable Relational Way’, which proposes a novel machine learning framework leveraging relational data representation and a selective naive Bayesian classifier. The approach transforms longitudinal patient data into interpretable features-derived via propositionalisation and informed by Minimum Description Length (MDL) principles-facilitating both accurate prediction and nuanced understanding of sepsis trajectories. Could this relational modeling paradigm unlock new avenues for personalized sepsis management and improved patient outcomes?
The Inevitable Cascade: Recognizing Sepsis in Time
Sepsis poses a formidable challenge within critical care medicine, consistently ranking as a leading cause of mortality and long-term morbidity in hospitals globally. This life-threatening condition, triggered by a dysregulated host response to infection, rapidly progresses beyond localized infection and necessitates immediate intervention. The complexity arises from sepsis’s often-subtle initial presentation, mimicking other common illnesses, and its ability to induce a cascade of physiological dysfunction affecting multiple organ systems. Consequently, healthcare providers face a constant imperative to swiftly recognize the early warning signs, differentiating sepsis from less critical conditions, and initiate appropriate treatment protocols – a task complicated by the heterogeneity of patient responses and the lack of a single, definitive diagnostic marker. Improved recognition and accelerated response times are, therefore, paramount in mitigating the devastating consequences of this widespread critical illness.
The consequences of delayed sepsis diagnosis are profound, directly correlating with a substantial increase in both the severity of illness – morbidity – and the risk of death – mortality. Each hour of delay in initiating appropriate treatment demonstrably worsens patient outcomes, as the systemic inflammatory response spirals and organ dysfunction accelerates. This critical time sensitivity underscores the pressing need for innovative detection methods that move beyond reliance on traditional, often sluggish, laboratory tests and subjective clinical assessments. Faster, more accurate diagnostic tools are not merely desirable, but essential to curtail the devastating impact of sepsis and improve survival rates for affected individuals, demanding a paradigm shift in how this life-threatening condition is identified and addressed.
Current sepsis diagnosis frequently hinges on clinical assessments – evaluating a patient’s temperature, heart rate, and mental status – which, while essential, are inherently subjective and prone to individual interpretation. This reliance on initial observations is often compounded by the necessary wait for definitive laboratory confirmation, such as blood cultures and complete blood counts, results of which can take 24 to 72 hours to materialize. This temporal disconnect creates a dangerous diagnostic delay, as sepsis progresses rapidly; each hour without appropriate antibiotic treatment demonstrably increases the risk of organ failure and death. Consequently, a critical gap exists between clinical suspicion and confirmed diagnosis, necessitating the development of more rapid and objective tools to facilitate earlier intervention and improve patient outcomes.
Data as Foundation: Mining the MIMIC-III Database
The MIMIC-III database, comprising data from Beth Israel Deaconess Medical Center, offers a publicly available and extensively documented collection of over 60,000 intensive care unit patient records. These records include demographics, vital signs, laboratory results, medications, and diagnostic codes, recorded over a ten-year period. The database is de-identified to protect patient privacy while maintaining the clinical integrity necessary for robust research. Its size and detailed nature make it a valuable resource for developing and validating machine learning models intended for clinical prediction and decision support, and it has been widely adopted by the research community for tasks such as predicting mortality, length of stay, and readmission rates.
Feature engineering within the MIMIC-III database begins with propositionalization, a method of transforming relational data into a format suitable for machine learning algorithms. This involves converting data stored across multiple tables and linked by relationships into a set of discrete, binary features. By representing each relationship and variable as a proposition – a statement that can be true or false – the data becomes amenable to analysis by algorithms requiring flat, tabular inputs. This process is a prerequisite for subsequent feature aggregation, enabling the creation of composite variables and ultimately improving model performance through enhanced data representation.
Feature aggregation is a key component of the data preparation pipeline, combining multiple individual variables into a smaller set of more informative features for machine learning models. This process facilitates robust variable selection by reducing dimensionality and potentially capturing complex relationships between original variables. Empirical results demonstrate optimal model performance is achieved utilizing between 98 and 1000 aggregated variables, suggesting a balance must be struck between retaining sufficient information and avoiding overfitting or computational inefficiency. The specific aggregation methods employed are not defined, but the observed performance range indicates that the technique effectively consolidates data for improved model training.
Missing data is a significant concern when utilizing the MIMIC-III database for machine learning model development, necessitating the application of data imputation techniques. Common methods include mean, median, and mode imputation, alongside more sophisticated approaches like k-nearest neighbors imputation. However, many healthcare datasets, including MIMIC-III, exhibit class imbalance, where the number of instances in one class significantly outweighs others. To mitigate the effects of both missing data and imbalance, imputation is often combined with the Synthetic Minority Oversampling Technique (SMOTE). SMOTE generates synthetic examples for the minority class by interpolating between existing minority class instances, effectively balancing the dataset and improving model performance, particularly for under-represented conditions.
Predictive Models: Mapping the Trajectory of Sepsis
Multiple machine learning algorithms are applicable to the prediction of sepsis onset, each with distinct characteristics. Logistic Regression provides a statistically interpretable model for estimating the probability of sepsis, while Random Forests, an ensemble method, leverage multiple decision trees to improve predictive accuracy and reduce overfitting. Gradient Boosting further refines this approach by sequentially building trees, weighting them based on their performance, and focusing on previously misclassified instances. These models utilize patient data, including vital signs, laboratory results, and demographics, as input features to identify patterns indicative of developing sepsis and generate predictions regarding the likelihood of its occurrence.
Time-series classification techniques are particularly relevant to sepsis prediction due to the disease’s progression over time, often manifesting as subtle changes in physiological signals. These methods analyze sequences of data points – such as heart rate, respiratory rate, and blood pressure – to identify patterns indicative of developing sepsis. The integration of attention mechanisms further refines this analysis by allowing the model to focus on the most salient time steps within the sequence, effectively weighting the importance of different data points in the prediction process. This targeted approach enables the model to better discern early warning signs and improve the accuracy of sepsis onset prediction compared to methods that treat all time points equally.
Bayesian Explainable Models offer a pathway to sepsis prediction systems prioritizing interpretability and trustworthiness through the application of Fractional Naive Bayes. This approach contrasts with ‘black box’ models by explicitly quantifying prediction uncertainty and providing probabilistic reasoning for each prediction. Fractional Naive Bayes allows for the modeling of feature dependencies, improving accuracy over traditional Naive Bayes while maintaining computational efficiency. The resulting models output posterior probabilities, enabling clinicians to assess the likelihood of sepsis and understand the contributing factors driving the prediction, facilitating informed decision-making and building confidence in the system’s recommendations. This transparency is crucial for clinical adoption and responsible implementation of predictive analytics in healthcare settings.
Evaluations of the machine learning models for sepsis prediction demonstrate strong performance, achieving an Area Under the Curve (AUC) ranging from 0.83 to 0.91. This level of accuracy is comparable to that of currently available commercial sepsis prediction systems. Critically, model performance is also supported by a defined prediction horizon, which represents the lead time – measured in hours – available for clinicians to implement proactive interventions following a positive sepsis prediction. The length of this prediction horizon is a key determinant of the model’s clinical utility and potential to improve patient outcomes.
Beyond Prediction: Trust, Transparency, and Timely Response
Bayesian Explainable Models are proving crucial in translating the complex outputs of sepsis prediction algorithms into clinically meaningful insights. These models don’t simply offer a probability of sepsis; they articulate the reasons behind that prediction, quantifying the contribution of each relevant factor – such as heart rate variability, lactate levels, or white blood cell count – to the overall risk assessment. This transparency is paramount for building clinician trust, as it allows healthcare professionals to evaluate the model’s logic, compare it to their own clinical judgment, and confidently integrate the predictions into patient care. By moving beyond ‘black box’ predictions, these models empower clinicians to understand why a patient is flagged as high-risk, facilitating more informed decision-making and ultimately, improved patient outcomes.
Counterfactual reasoning offers a powerful lens through which to understand the rationale behind complex predictive models, particularly in critical care settings. Rather than simply identifying that a patient is at high risk for sepsis, these techniques delve into what specific changes in a patient’s condition would have altered the prediction. For example, a counterfactual analysis might reveal that a slight decrease in heart rate, or a different white blood cell count, would have shifted a patient from a high-risk to a low-risk category. This capability moves beyond mere prediction, providing clinicians with actionable insights – a ‘what if’ scenario – to better understand the driving forces behind the model’s assessment and potentially inform clinical decision-making. By pinpointing these pivotal factors, counterfactual explanations enhance trust in the model and facilitate a more nuanced interpretation of its predictions, ultimately supporting more informed and proactive patient care.
The capacity to accurately forecast sepsis, demonstrated by achieved Area Under the Curve (AUC) scores ranging from 0.83 to 0.91, offers a critical window for timely medical response. This predictive power isn’t merely statistical; it directly translates to the potential for earlier interventions, such as the prompt administration of antibiotics or fluid resuscitation. Sepsis, a life-threatening condition arising from the body’s overwhelming response to infection, demands rapid diagnosis and treatment to prevent organ dysfunction and reduce the risk of mortality. Consequently, a model capable of identifying patients at high risk with such precision enables clinicians to initiate potentially life-saving therapies sooner, ultimately contributing to decreased morbidity and improved patient survival rates in the critical care setting.
The convergence of interpretable machine learning and critical care medicine promises a shift towards predictive, individualized treatment strategies. Rather than reacting to established sepsis, these advancements enable clinicians to anticipate risk and tailor interventions based on a patient’s unique physiological trajectory. This proactive approach extends beyond simply flagging high-risk individuals; counterfactual analyses reveal the specific factors influencing a prediction, allowing for targeted adjustments to care plans. Consequently, early and precise interventions become feasible, potentially mitigating the organ dysfunction and mortality rates historically associated with sepsis, and ultimately fostering improved outcomes through a more personalized and preventative paradigm of critical care.
The pursuit of predictive modeling, as demonstrated in this temporal sepsis analysis, inevitably confronts the realities of system decay. Just as structures erode over time, predictive models require constant refinement to maintain accuracy against shifting data landscapes. Alan Turing observed, “No system is immune to the possibility of error.” This sentiment resonates deeply with the article’s emphasis on relational data and Bayesian classification; these methods aren’t about achieving perfect prediction, but about constructing a system that gracefully degrades, offering interpretable insights even as its predictive power diminishes. The model’s focus on Minimum Description Length (MDL) acknowledges this inherent entropy, seeking the simplest explanation that best fits the observed data, a strategy for resisting the inevitable accumulation of ‘technical debt’ within the system.
What Lies Ahead?
The pursuit of early sepsis detection, as demonstrated by this work, inevitably confronts the limitations inherent in predictive modeling itself. Accuracy, while valuable, is a transient metric. The true test resides in a model’s graceful degradation as the underlying patient population shifts, or as healthcare practices evolve. Each abstraction-each carefully selected feature-carries the weight of the past, potentially obscuring emergent signals of change. Future investigations must focus not solely on improving predictive power, but on quantifying and mitigating the rate of obsolescence.
The relational approach, while promising for its interpretability, introduces its own temporal challenges. Maintaining a coherent and useful representation of patient relationships over time requires ongoing refinement. The Minimum Description Length principle, employed here for feature selection, represents a pragmatic compromise between model complexity and accuracy. However, a more nuanced understanding of information loss during model simplification is critical.
Ultimately, the most resilient systems are those that embrace slow change. The field should explore methods for continuous model adaptation, perhaps through incremental learning or meta-modeling techniques. The goal isn’t to prevent decay, but to manage it-to build systems that learn from their own obsolescence and adapt accordingly.
Original article: https://arxiv.org/pdf/2601.21747.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- All Golden Ball Locations in Yakuza Kiwami 3 & Dark Ties
- All Itzaland Animal Locations in Infinity Nikki
- NBA 2K26 Season 5 Adds College Themed Content
- Critics Say Five Nights at Freddy’s 2 Is a Clunker
- Super Animal Royale: All Mole Transportation Network Locations Guide
- Gold Rate Forecast
- Brent Oil Forecast
- BREAKING: Paramount Counters Netflix With $108B Hostile Takeover Bid for Warner Bros. Discovery
- Unlocking the Jaunty Bundle in Nightingale: What You Need to Know!
- Elder Scrolls 6 Has to Overcome an RPG Problem That Bethesda Has Made With Recent Games
2026-01-31 06:03