Author: Denis Avetisyan
A new deep learning framework leverages the power of Kolmogorov-Arnold Networks to predict species co-occurrence and improve joint species distribution models.

LabelKAN utilizes inter-label learning to enhance both the accuracy and interpretability of ecological predictions based on species relationships.
Despite growing efforts to reverse global biodiversity loss, accurately modeling species distributions and ecological relationships remains a significant challenge. This is addressed in ‘LabelKAN — Kolmogorov-Arnold Networks for Inter-Label Learning: Avian Community Learning’, which introduces a novel framework leveraging Kolmogorov-Arnold Networks to improve joint species distribution models by explicitly learning species co-occurrence patterns. Our approach demonstrates substantial gains in predictive performance, particularly for rare and difficult-to-predict species, and enhances confidence in both species distributions and community structure predictions. Will this inter-label learning approach unlock a new generation of ecological models capable of more effectively informing conservation strategies?
Emergent Patterns in Species Distribution
Predicting where species live demands a deep understanding of the connections between organisms and their environment, yet this pursuit is consistently challenged by incomplete information. Species aren’t distributed randomly; their presence is dictated by a complex interplay of factors like climate, topography, vegetation, and the presence of other species – relationships that are difficult to fully capture. Crucially, robust predictions require extensive data on species occurrences, but such data is often sparse, unevenly distributed, or simply unavailable, especially for elusive or rare organisms. This limitation forces scientists to rely on statistical inferences and modeling techniques, acknowledging inherent uncertainties in mapping the distributions of life across the globe. Consequently, projections of species ranges must be interpreted with caution, as they represent informed estimates rather than definitive depictions of reality.
Conventional Species Distribution Models (SDMs) often fall short when predicting where species actually live, primarily because these models typically treat each species in isolation. This approach overlooks the complex web of interactions – competition, predation, symbiosis – that fundamentally shape species’ ranges. Consequently, SDMs struggle to accurately predict species co-occurrence; a model predicting suitable habitat for species A might not align with the observed presence of species B, even if the two are tightly linked ecologically. Furthermore, modeling rare species presents a unique challenge, as the limited data available can lead to inaccurate estimations of habitat suitability and inflated uncertainty; traditional SDMs require sufficient presence data to reliably delineate a species’ niche, a requirement often unmet for species with naturally small populations or patchy distributions. This necessitates the development of more sophisticated modeling techniques capable of accounting for interspecific relationships and effectively handling sparse data to improve predictive accuracy and inform conservation efforts.
The surge in biodiversity data collected by citizen scientists through platforms like iNaturalist and eBird presents a transformative opportunity for ecological forecasting. These initiatives generate datasets of a scale previously unattainable, offering insights into species distributions and phenology across vast geographical areas and extended timeframes. However, this wealth of information is not without its challenges. Data collected by untrained observers can be subject to biases – certain species or locations may be over-represented due to observer effort, taxonomic expertise, or accessibility. Furthermore, issues of data quality, including misidentification and spatial inaccuracies, require careful consideration. Researchers are actively developing statistical methods and validation techniques to account for these biases, ensuring that citizen science data can be effectively integrated into robust ecological models and ultimately improve the accuracy of predictions about the natural world.
Deep Learning: Unveiling Latent Ecological Structures
Deep learning techniques facilitate the modeling of intricate ecological relationships by leveraging latent space representations, which are lower-dimensional abstractions of high-dimensional ecological data. These representations allow algorithms to identify and quantify underlying factors driving species distributions and community composition without explicitly modeling every variable. By mapping ecological data into a latent space, deep learning models can capture non-linear relationships and complex interactions between species and their environment more efficiently than traditional statistical methods. This approach is particularly useful when dealing with large datasets containing numerous species and environmental predictors, as it reduces computational demands and mitigates the curse of dimensionality, ultimately improving the accuracy and interpretability of ecological models.
Variational Autoencoders (VAEs) and Deep Multivariate Probit Models (DMPMs) address challenges inherent in modeling species-environment relationships by reducing dimensionality and improving parameterization. VAEs accomplish this through non-linear dimensionality reduction, learning a compressed, latent space representation of ecological data which captures key environmental gradients and species occurrences. DMPMs, conversely, utilize a probabilistic framework to model the co-occurrence of multiple species given environmental covariates; deep learning architectures within DMPMs allow for flexible modeling of non-linear relationships and efficient estimation of parameters, particularly in high-dimensional datasets where traditional methods become computationally intractable. Both approaches enable researchers to move beyond simple linear models and capture complex ecological processes while mitigating overfitting and improving predictive performance.
Deep Neural Networks (DNNs) form the foundational architecture for many deep learning applications in ecology, facilitating the identification of non-linear relationships within complex datasets. These networks consist of multiple layers of interconnected nodes, allowing them to learn hierarchical representations of ecological variables. By processing data through these layers, DNNs can automatically extract features and patterns that may be obscured through traditional statistical methods. This is particularly useful in ecological modeling, where interactions between species and environmental factors are often intricate and high-dimensional. The ability of DNNs to handle large datasets and discover subtle relationships makes them a valuable tool for analyzing ecological data and generating predictive models.
LabelKAN: A Framework for Interconnected Species Modeling
LabelKAN integrates Deep Neural Networks (DNNs) and Kolmogorov-Arnold Networks (KANs) to improve predictive performance through enhanced inter-label learning. DNNs excel at feature extraction and complex pattern recognition, while KANs provide a mechanism for modeling relationships between labels – in this context, species – through their inherent network structure. This combination allows LabelKAN to not only identify individual species characteristics but also to learn how species co-occur and influence each other’s distributions. The KAN component facilitates the sharing of information between species labels during training, leading to a more robust and accurate model capable of improved prediction, particularly for species with limited data.
Kolmogorov-Arnold Networks (KANs) contribute to LabelKAN’s enhanced modeling capability by representing functions as sums of radial basis functions, allowing for a more flexible and expressive representation of species relationships than traditional deep neural networks. This approach enables the model to capture non-linear interactions and dependencies between species and environmental variables with greater precision. Specifically, KANs facilitate the identification of complex co-occurrence patterns and subtle environmental influences that impact species distributions, improving the model’s ability to generalize to unseen data and accurately predict the presence or absence of species across varying landscapes and conditions. This is particularly beneficial for modeling species interactions where relationships are not strictly linear or additive.
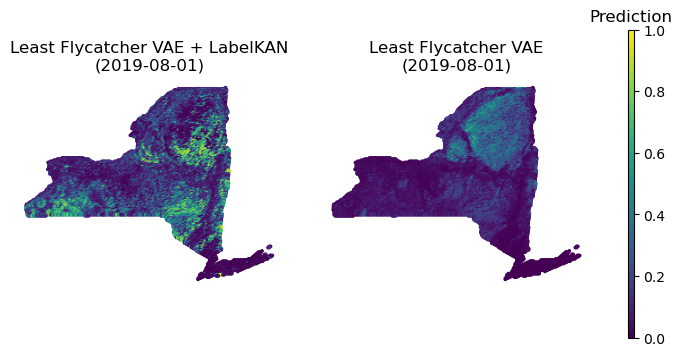
LabelKAN addresses the complexities of species co-occurrence modeling, a significant challenge in ecological forecasting due to interspecies relationships and varying detection probabilities. Traditional methods often struggle with accurately predicting distributions, particularly for rare species where data is limited. LabelKAN improves prediction accuracy for both common and rare species by leveraging inter-label learning; the framework learns relationships between species, allowing it to infer the presence of one species based on the known distribution of others. This approach effectively mitigates the impact of sparse data for rare species, leading to more robust and reliable distribution predictions across all species within the modeled community.
Evaluation of the LabelKAN framework using a case study focused on the Great Blue Heron demonstrated its practical application to ecological forecasting. Results indicated a measurable improvement in predictive performance, with the model achieving a 2-3% increase in both micro and macro-aggregated Area Under the Precision-Recall Curve (AUPRC). This improvement is attributed to LabelKAN’s ability to effectively model species co-occurrence patterns, leading to more accurate predictions of species distributions.

Towards Transparent and Actionable Ecological Insights
LabelKAN distinguishes itself not merely through heightened predictive performance, but also by revealing why a model arrives at a specific prediction, thereby unlocking crucial insights into species-environment relationships. Traditional ecological forecasting often operates as a ‘black box’, providing predictions without elucidating the underlying ecological drivers; LabelKAN overcomes this limitation by identifying the specific environmental variables most influential in determining species distributions. This enhanced interpretability allows researchers to move beyond simply knowing where a species is likely to be found, to understanding how and why it responds to particular environmental conditions. Consequently, LabelKAN fosters a more nuanced understanding of ecological processes, enabling more informed conservation planning and adaptive management strategies, and offering a pathway to validate and refine existing ecological theory.
LabelKAN distinguishes itself through a unique capacity to integrate disparate data types in ecological modeling. Traditionally, species distribution models rely heavily on confirmed presence and absence data, which can be costly and time-consuming to obtain. However, citizen science initiatives like iNaturalist generate vast quantities of presence-only records – observations of where a species is found, without systematically surveying where it is not. LabelKAN skillfully combines these readily available presence-only data with more traditional presence-absence datasets, creating a more comprehensive and robust picture of species distributions. This flexibility not only expands the scope of possible analyses, particularly in data-poor regions, but also enhances the reliability of predictions by accounting for biases inherent in different data sources, ultimately leading to more actionable insights for ecological forecasting.
Accurate modeling of rare species presents a significant challenge in ecological forecasting, yet is paramount for effective conservation. This framework addresses this need by employing a novel approach to data integration and statistical modeling, enabling reliable predictions even with limited occurrences. The ability to pinpoint suitable habitats and anticipate population shifts for these vulnerable species directly informs targeted management strategies – from prioritizing conservation areas and allocating resources to mitigating threats and monitoring recovery. By moving beyond broad-scale predictions, conservationists can now focus efforts on the specific locations and interventions most likely to benefit species at greatest risk, improving the efficiency and impact of conservation initiatives and bolstering long-term biodiversity.
The LabelKAN framework, initially demonstrated with select species, holds considerable promise for widespread implementation across a variety of ecological contexts. Its adaptability to both presence-absence and presence-only data streams – including citizen science initiatives – circumvents a common limitation in species distribution modeling, allowing for predictions even with incomplete datasets. This scalability is particularly valuable given the accelerating rate of environmental change and the urgent need for proactive conservation strategies; by facilitating accurate forecasts of species responses to shifting conditions, LabelKAN offers a pathway towards more informed and effective resource management, ultimately bolstering resilience within increasingly vulnerable ecosystems. The framework’s potential extends beyond single-species predictions, paving the way for holistic assessments of biodiversity and community-level responses to disturbance.
The research detailed in this paper demonstrates a shift from attempting to centrally control ecological understanding-dictating species relationships through pre-defined models-to allowing patterns to emerge from the data itself. This aligns with the observation that “knowledge is power,” as Francis Bacon posited. LabelKAN, by leveraging Kolmogorov-Arnold Networks, doesn’t impose a singular structure but instead facilitates the discovery of inherent co-occurrence patterns within avian communities. The framework acknowledges the complexity of ecological systems and recognizes that attempting to override natural order with rigid models limits both predictive accuracy and genuine insight into species interactions. It’s a system where local rules – the relationships between species – produce global effects, mirroring the emergent behavior the study elucidates.
What Lies Ahead?
The pursuit of predictive accuracy in joint species distribution models, as exemplified by LabelKAN, often feels like an attempt to impose order upon inherently stochastic systems. The framework demonstrates an ability to capture co-occurrence, but it is crucial to remember that these patterns emerge; they are not dictated by any underlying ecological blueprint. Future work should prioritize understanding the limits of predictability, rather than striving for ever-more-complex models that chase diminishing returns.
The strength of LabelKAN resides in its network architecture, and the implicit learning of relationships. However, interpretability, while improved, remains a challenge. A shift in focus toward quantifying influence – how one species subtly alters the probabilities of others – may prove more fruitful than seeking definitive causal links. Robustness emerges from these decentralized interactions; it cannot be designed through centralized control.
Ultimately, the field would benefit from acknowledging that complete knowledge of an ecological system is an illusion. Instead of attempting to build a perfect model, the emphasis should be on developing frameworks that can adapt and evolve alongside the systems they attempt to represent. System structure is stronger than individual control, and future research should reflect that principle.
Original article: https://arxiv.org/pdf/2601.18818.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- United Airlines can now kick passengers off flights and ban them for not using headphones
- All Golden Ball Locations in Yakuza Kiwami 3 & Dark Ties
- How To Find All Jade Gate Pass Cat Play Locations In Where Winds Meet
- How to Complete Bloom of Tranquility Challenge in Infinity Nikki
- Every Battlefield game ranked from worst to best, including Battlefield 6
- Best Zombie Movies (October 2025)
- Gold Rate Forecast
- 29 Years Later, A New Pokémon Revival Is Officially Revealed
- How School Spirits Season 3 Ending Twist Will Impact Season 4 Addressed By Creators
- The ARC Raiders Dev Console Exploit Explained
2026-01-28 22:34