Author: Denis Avetisyan
New research dives into the ‘black box’ of deep learning models to understand why they predict lung cancer risk, going beyond simply measuring if they are correct.
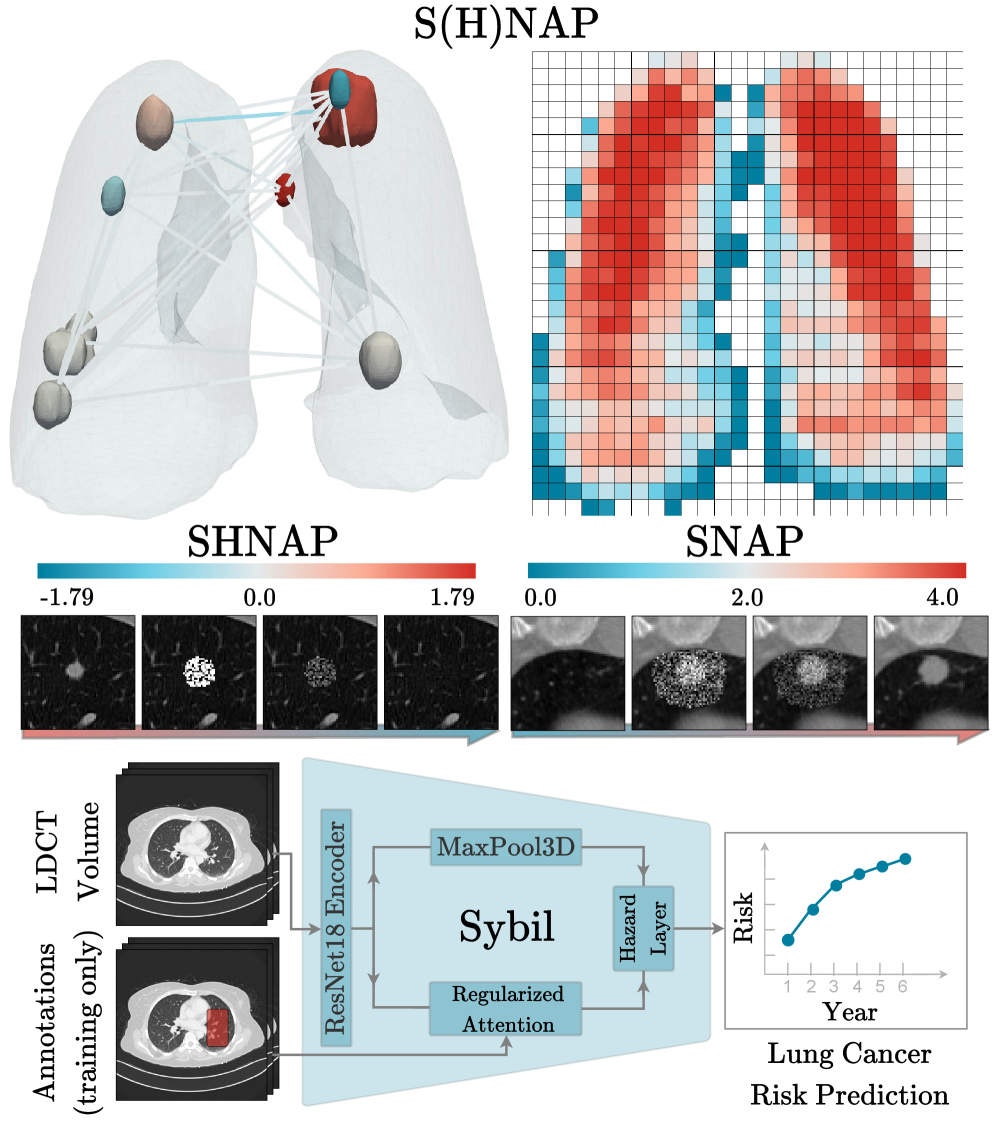
This study introduces SHNAP and SNAP, novel interventional auditing methods to explain deep learning predictions for lung cancer risk assessment.
Despite advances in automated screening, the clinical deployment of deep learning models for lung cancer risk prediction requires more than just high accuracy. This work, ‘Auditing Sybil: Explaining Deep Lung Cancer Risk Prediction Through Generative Interventional Attributions’, introduces S(H)NAP, a model-agnostic auditing framework that moves beyond observational metrics to reveal the causal reasoning of the Sybil deep learning model. Through generative interventional attributions, we demonstrate that while Sybil often mimics expert radiological assessment, it exhibits critical failure modes-including sensitivity to irrelevant artifacts and a radial bias-that could impact clinical reliability. Can a truly robust and trustworthy AI for lung cancer screening be achieved without a comprehensive understanding of how these models arrive at their conclusions?
The Challenge of Distinguishing Benign from Malignant Nodules
Lung cancer screening with Low-Dose Computed Tomography (LDCT) frequently reveals pulmonary nodules-small spots in the lung-but a significant challenge lies in differentiating benign from malignant growths. This leads to a high rate of false positive results, where patients are flagged as potentially having cancer when they do not. Consequently, numerous individuals undergo further, often invasive, diagnostic procedures-like biopsies-that are ultimately unnecessary and carry their own risks. These interventions not only cause patient anxiety and discomfort but also strain healthcare resources, highlighting the critical need for improved accuracy in initial nodule assessments to minimize overdiagnosis and ensure effective lung cancer screening programs.
Lung cancer screening with low-dose computed tomography often reveals a high number of pulmonary nodules, presenting a significant challenge in distinguishing between benign and malignant growths. Accurate risk stratification is crucial, yet complicated by the subtle nuances in nodule characteristics – size, shape, density, and texture – as well as their specific location within the lungs. Even minor variations in these features can influence the likelihood of malignancy, demanding sophisticated analytical approaches. The sheer volume of nodules necessitates efficient and precise evaluation; a process hindered by the difficulty in capturing the complex interplay between these characteristics and their combined impact on predicting cancerous development. Consequently, refining the ability to accurately assess individual nodule risk is paramount to reducing false positives and ensuring appropriate clinical follow-up.
Current approaches to lung cancer risk prediction often fall short due to an inability to fully account for the nuanced relationships between various nodule characteristics and malignancy. While individual features – such as size, shape, and texture – offer some predictive value, it is the combination of these features, and their complex interactions, that ultimately determines a nodule’s likelihood of being cancerous. Existing computational models frequently treat these features in isolation or rely on simplified assumptions about their interplay, thereby overlooking critical information. This limitation leads to imprecise predictions, increased false positive rates, and ultimately, hinders the effectiveness of early detection programs. A more sophisticated understanding of these feature interactions is therefore crucial for developing models capable of accurately stratifying risk and improving patient outcomes.
Accurate early detection of lung cancer hinges on effectively evaluating the risk posed by pulmonary nodules identified in screening scans, but current predictive models often operate as ‘black boxes’. Recent work has focused on dissecting the logic of complex models, such as Sybil, to better understand how risk is assessed. This research demonstrates a framework capable of approximating Sybil’s predictions using a surprisingly simple linear model that accounts for interactions between pairs of nodule characteristics. Achieving an R² value of approximately 1 signifies that this linear approximation faithfully replicates Sybil’s behavior, offering a transparent and interpretable alternative for risk stratification and potentially reducing unnecessary follow-up procedures. This ability to distill complex algorithms into understandable components is a crucial step towards improving the precision and effectiveness of lung cancer screening programs.

Deconstructing Sybil: A Nodule-Centric Analysis
Sybil is a deep learning model designed to assess lung cancer risk utilizing computed tomography (CT) scan data. While demonstrating predictive capability, the internal mechanisms driving Sybil’s risk assessments are not inherently transparent; the model operates as a complex, non-linear function making it difficult to discern why a specific prediction was made. This lack of interpretability is a common challenge with deep learning models, hindering clinical trust and the ability to validate the model’s reasoning against established radiological principles. Consequently, understanding the basis for Sybil’s predictions is crucial for both improving the model and facilitating its responsible deployment in clinical settings.
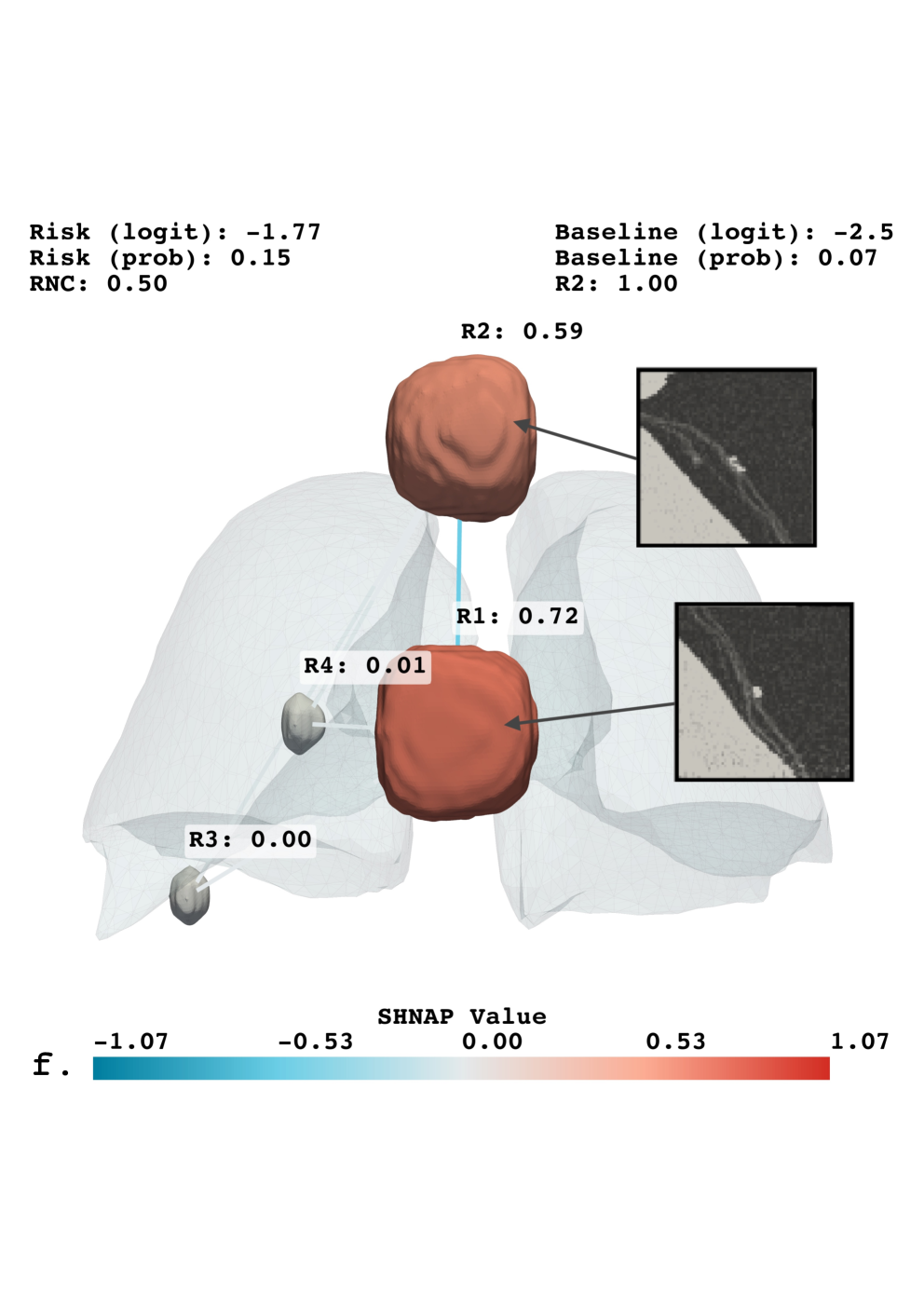
SHNAP, or SHapley values for Nodules and Pairwise interactions, is a decomposition framework used to analyze the predictions made by the Sybil lung cancer risk assessment model. It operates by approximating Sybil’s complex decision function with a linear model that explicitly accounts for the contributions of individual lung nodules and the interactions between them. This approximation enables the calculation of Shapley values, a concept from game theory, which quantifies the marginal contribution of each nodule and nodule pair to the overall prediction. By attributing predictive power in this manner, SHNAP provides a granular, interpretable explanation of Sybil’s reasoning process, revealing which nodules are most influential in driving the model’s risk assessment for a given CT scan.
The methodology employed to interpret Sybil’s predictions utilizes a Linear Model with Pairwise Interactions as a surrogate for its complex, non-linear decision function. This approximation allows for the decomposition of Sybil’s output into additive contributions from individual nodules and their interactions; specifically, the model estimates a weight for each nodule and each nodule pair representing their influence on the final risk score. The linearity of this model is crucial, as it enables direct attribution of feature importance through the learned weights, facilitating interpretable explanations of Sybil’s predictions based on quantifiable nodule characteristics and relationships. The model’s form is as follows: f(x) = w_0 + \sum_{i=1}^{N} w_i x_i + \sum_{i=1}^{N} \sum_{j=i+1}^{N} w_{ij} x_i x_j , where x_i represents the feature vector for nodule i, and w terms represent the learned weights.
Quantification of individual nodule influence on Sybil’s lung cancer risk predictions allows for the identification of salient features driving its assessments. The SHNAP framework achieves this by decomposing Sybil’s output and attributing contributions to each nodule and their pairwise interactions. Critically, the stability of these explanations across the diverse iLDCT dataset is demonstrated by low standard deviation values observed in the SHNAP attribution scores; this indicates consistent feature importance across different patients and imaging characteristics, suggesting robustness in Sybil’s reasoning process and reliable interpretability of its predictions.
Generating Synthetic Scenarios for Rigorous Model Validation
Schrödinger Bridge Diffusion (SDB) is a generative model employed for the synthetic manipulation of Computed Tomography (CT) scans, specifically enabling the realistic removal or insertion of pulmonary nodules. This is achieved through a diffusion process that learns the underlying distribution of nodule characteristics within CT imagery, allowing for the creation of modified scans that maintain anatomical plausibility. The model operates by iteratively adding or removing noise to the image data, guided by a bridging function that ensures coherence between the original and modified scans. SDB facilitates controlled experimentation by providing a mechanism to precisely alter nodule features – such as size, shape, and texture – and their spatial location within the lung volume, without introducing artifacts that would invalidate the evaluation process.
Sybil’s spatial sensitivity is assessed through datasets generated using Schrödinger Bridge Diffusion (SDB) in combination with the SHNAP and SNAP frameworks. SHNAP provides precise control over nodule placement and characteristics within Computed Tomography (CT) scans, while SNAP facilitates the automated generation of numerous synthetic scans with varied nodule configurations. SDB then leverages these configurations to create realistic CT volumes incorporating the specified nodules, effectively allowing for systematic variation of nodule size, shape, location, and surrounding tissue characteristics. This combined approach enables the creation of diverse and controlled datasets specifically designed to probe Sybil’s performance across a range of spatial arrangements and subtle anatomical variations, exceeding the limitations of purely relying on existing datasets.
Systematic variation of nodule characteristics – including size, shape, texture, and density – coupled with controlled manipulation of nodule location within the CT scan volume, allows for a targeted assessment of Sybil’s sensitivity to subtle changes in pulmonary nodule presentation. This process facilitates the identification of potential biases in Sybil’s detection or classification performance; for example, whether the model exhibits decreased accuracy with nodules of a specific size range or located in particular anatomical regions. By quantifying performance across this range of synthetic data, we can determine the limits of Sybil’s robustness and areas requiring further refinement to ensure consistent and reliable nodule analysis.
The synthetic datasets generated via Schrödinger Bridge Diffusion (SDB) are utilized alongside established benchmarks, LUNA25 and iLDCT, to provide a more comprehensive and robust evaluation of Sybil’s performance. Quantitative analysis demonstrates SDB’s superior reconstruction quality compared to other generative methods; specifically, SDB achieves a lower Fréchet Inception Distance (FID) – a metric indicating greater similarity to real CT scans – than baseline methods, as detailed in Table 1. This improved fidelity of the generated data allows for more rigorous probing of Sybil’s capabilities and potential biases in nodule detection and characterization.
Unveiling Spatial Bias and the Power of Counterfactual Reasoning
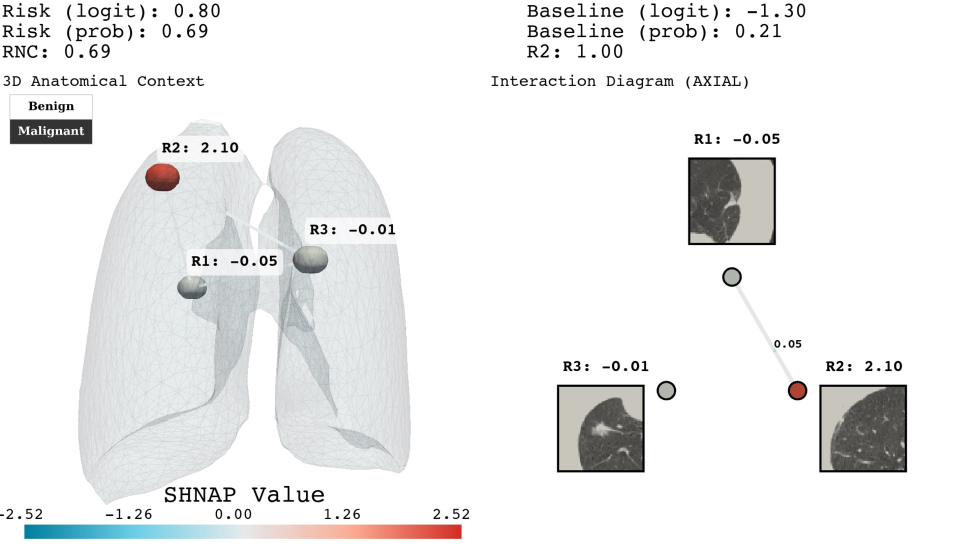
Analysis of Sybil, an AI system for lung nodule malignancy assessment, reveals a consistent underestimation of risk for nodules located closer to the pleura – the lung’s outer lining. This phenomenon, termed Radial Sensitivity Bias, indicates that the system systematically assigns lower malignancy probabilities to nodules as their proximity to the pleura increases. The bias isn’t random; it’s a patterned decrease in sensitivity directly correlated with radial distance from the pleura, suggesting the model doesn’t adequately account for the impact of this anatomical location on the likelihood of malignancy. Consequently, clinically relevant nodules near the pleura may be overlooked, potentially delaying crucial interventions and impacting patient outcomes. This finding underscores the critical need to address such spatial biases in AI-driven diagnostic tools.
The research team developed SNAP, a spatial probing framework, to dissect the underlying causes of risk underestimation in lung nodule assessments. By systematically manipulating the spatial context of nodules within imaging data and leveraging counterfactual explanations, SNAP identifies the specific features driving the model’s predictions. This approach doesn’t simply highlight that a bias exists, but rather elucidates why the AI assigns lower risk to nodules near the pleura. Counterfactual analysis, in particular, reveals how subtly altering a nodule’s position – moving it away from the pleural surface – dramatically shifts the predicted malignancy score. The combination of spatial probing and counterfactual reasoning provides a granular understanding of the model’s decision-making process, pinpointing the critical interplay between nodule location and AI assessment.
Investigations into Sybil, an AI designed for lung nodule malignancy assessment, reveal limitations in how its attention mechanism processes spatial information. The system doesn’t fully integrate the clinical significance of a nodule’s proximity to the pleura – the lung’s outer lining – when determining risk. While Sybil effectively identifies salient features within the nodule itself, its attentional weighting appears insufficient to capture the increased likelihood of malignancy often associated with pleural proximity, a factor routinely considered by radiologists. This suggests that the model prioritizes intrinsic nodule characteristics over crucial contextual cues, potentially leading to underestimation of risk for nodules located near the pleura and highlighting a need for enhanced spatial reasoning capabilities in AI-driven diagnostic tools.
The consistent underestimation of lung nodule malignancy risk in areas close to the pleura presents a critical challenge for AI-powered screening systems. Analysis utilizing the spatial probing framework, SNAP, reveals a systematic decrease in diagnostic sensitivity as nodule proximity to the pleura increases – a phenomenon termed Radial Sensitivity Bias. This isn’t simply a matter of image quality; the attribution trends pinpoint a failure of the AI’s attention mechanism to adequately account for the influence of pleural proximity. Consequently, even nodules with characteristics suggestive of malignancy may be incorrectly classified as benign when located near the pleura, underscoring the urgent need to address these spatial biases and refine AI algorithms for more accurate and reliable lung cancer screening.
The pursuit of demonstrable correctness, central to the paper’s introduction of SHNAP and SNAP, echoes a sentiment deeply held by mathematicians. G.H. Hardy famously stated, “A mathematician, like a painter or a poet, is a maker of patterns.” This crafting of patterns, in the context of deep learning auditing, isn’t merely about achieving high prediction accuracy for lung cancer risk. Instead, the paper emphasizes understanding why a model arrives at a specific conclusion. SHNAP and SNAP facilitate this understanding through interventional analysis, moving beyond superficial feature importance to reveal the causal mechanisms driving predictions – a rigorous demonstration of the ‘pattern’ the model employs, proving its logic beyond empirical observation. This aligns with the core principle that a provable solution, even if complex, is superior to one based solely on observed functionality.
What Lies Ahead?
The introduction of SHNAP and SNAP represents a necessary, if incremental, step toward verifiable intelligence in a domain of considerable consequence. Current methodologies, fixated on aggregate performance, offer little insight into the reasoning – or lack thereof – underlying a prediction. The assertion that a model achieves 90% accuracy is, mathematically speaking, vacuous without a corresponding understanding of the error landscape and the invariants governing correct classification. Future work must move beyond merely identifying influential features to formally characterizing the conditions under which a model’s output is demonstrably correct, ideally with provable guarantees.
A significant limitation remains the reliance on observational data. While SHNAP and SNAP refine the audit of existing models, they do not address the fundamental problem of spurious correlations inherent in observational studies. The true test of these interventional attribution methods will lie in their application to controlled experiments – data generated under conditions where causal relationships are known a priori. Only then can one definitively assess whether the identified attributions reflect genuine causal mechanisms, or merely statistical artifacts.
Ultimately, the pursuit of explainable AI is not about making black boxes appear transparent, but about constructing models that are intrinsically interpretable. The asymptotic ideal remains a system where prediction and explanation are unified – a model whose internal logic is as readily verifiable as a theorem in Euclidean geometry. Whether deep learning, in its current form, can achieve this remains an open – and perhaps, fundamentally challenging – question.
Original article: https://arxiv.org/pdf/2602.02560.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- All Golden Ball Locations in Yakuza Kiwami 3 & Dark Ties
- Gold Rate Forecast
- Hollywood is using “bounty hunters” to track AI companies misusing IP
- What time is the Single’s Inferno Season 5 reunion on Netflix?
- Mario Tennis Fever Review: Game, Set, Match
- NBA 2K26 Season 5 Adds College Themed Content
- A Knight Of The Seven Kingdoms Season 1 Finale Song: ‘Sixteen Tons’ Explained
- Train Dreams Is an Argument Against Complicity
- Every Death In The Night Agent Season 3 Explained
- 2026 Upcoming Games Release Schedule
2026-02-04 21:12