Author: Denis Avetisyan
A new hybrid AI framework, AgriPINN, combines the power of deep learning with established agricultural models for more accurate and interpretable crop biomass predictions under challenging conditions.
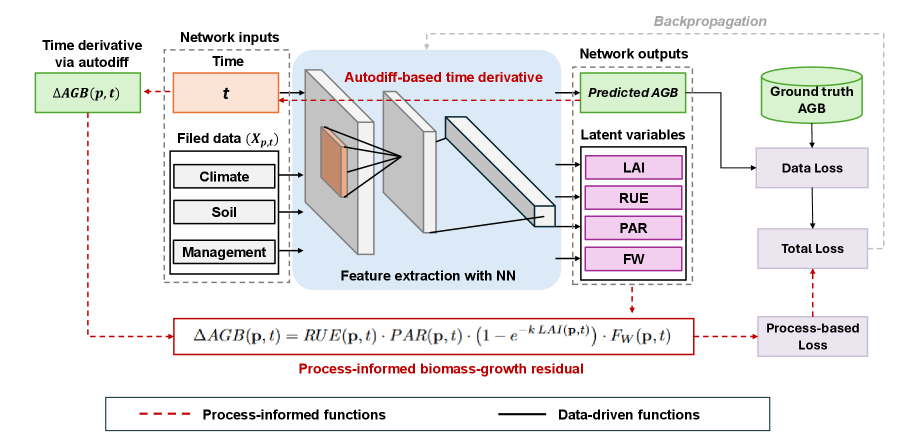
This work presents a process-informed neural network approach for scalable above-ground biomass estimation and water stress assessment in crops.
Accurate and scalable prediction of crop productivity under increasingly variable water availability remains a significant challenge for modern agriculture. This is addressed in ‘AgriPINN: A Process-Informed Neural Network for Interpretable and Scalable Crop Biomass Prediction Under Water Stress’, which introduces a novel hybrid modeling approach integrating biophysical principles into a deep learning framework. By embedding a crop-growth differential equation as a differentiable constraint, AgriPINN achieves improved accuracy and interpretability in predicting above-ground biomass while maintaining computational efficiency. Could this fusion of process-based knowledge and deep learning unlock new capabilities for climate-resilient agricultural management and resource planning?
The Persistent Challenge of Data Scarcity in Crop Modeling
The challenge of ensuring global food security is increasingly reliant on the ability to accurately predict crop yields, yet traditional process-based modeling approaches are frequently hampered by a critical lack of comprehensive data. These models, designed to simulate plant growth based on underlying physiological processes, demand extensive information regarding soil properties, weather patterns, and crop characteristics for effective parameterization. However, such detailed data is often unavailable, particularly in regions most vulnerable to food insecurity, creating significant uncertainty in predictions. This data scarcity isn’t merely a matter of incomplete datasets; it represents a fundamental bottleneck that limits the ability to translate complex biophysical understanding into actionable insights for agricultural management and policy decisions, hindering efforts to proactively address potential food supply disruptions.
Process-based crop models, such as LINTUL5, fundamentally simulate plant growth by explicitly representing underlying biophysical processes – photosynthesis, respiration, nutrient uptake, and the like. This approach allows them to accurately capture how crops respond to environmental constraints, like water stress or temperature extremes, offering a distinct advantage over purely empirical models. However, these models require numerous parameters – values that define specific plant traits or physiological rates – and accurately determining these parameters for diverse environments presents a significant challenge. While a model might perform well in the conditions used for calibration, its ability to generalize – to reliably predict crop behavior in novel locations or under different management practices – often diminishes due to the difficulty of capturing the full range of natural variability in plant characteristics and environmental interactions. This limitation highlights a core trade-off: the strength of process-based models lies in their mechanistic representation, but their practical utility is frequently constrained by the need for extensive, location-specific data for robust parameterization and validation.
The predictive capacity of process-based crop models faces a significant constraint due to widespread data scarcity, particularly when quantifying crucial variables like Above-ground Biomass (AGB). Accurate AGB estimation is vital for assessing crop yields, carbon sequestration potential, and overall ecosystem health, but obtaining sufficient ground-truth data for model calibration and validation remains a major challenge, especially in data-poor regions. This limitation hinders the ability of models to generalize across diverse environments and agricultural practices, leading to substantial uncertainties in predictions. Consequently, while these models offer a robust framework for understanding biophysical processes, their practical utility is diminished without comprehensive data to refine parameterization and ensure reliable outputs, ultimately impacting their effectiveness for informed decision-making in agriculture and land management.
A Synergistic Approach: Process-Informed Neural Networks
Process-Informed Neural Networks (PINNs) represent a hybrid modeling approach that combines the data-driven capabilities of artificial neural networks with existing domain knowledge expressed as biophysical constraints. This framework moves beyond traditional “black box” machine learning by directly incorporating governing equations or established models into the network’s learning process. Instead of solely relying on large datasets to infer relationships, PINNs utilize this pre-existing knowledge to guide the neural network’s parameter optimization, resulting in models that are more robust, require less training data, and are capable of generalizing to scenarios not explicitly represented in the training set. The integration is achieved by defining a loss function that penalizes deviations from the known biophysical behavior, effectively shaping the learned solution space.
The Process-Informed Neural Network (PINN) framework leverages the LINTUL5 model to establish a biophysical constraint during training. LINTUL5, a well-established plant and soil simulation model, generates synthetic training data representing expected biophysical relationships. This data isn’t used as the sole training set, but rather incorporated into the loss function as a ‘ProcessResidual’ term. By minimizing the difference between the neural network’s predictions and the LINTUL5 outputs, the PINN is actively guided toward solutions that adhere to known biophysical principles, effectively regularizing the learning process and improving the model’s ability to generalize beyond the observed data. This constraint ensures the neural network doesn’t produce outputs that violate fundamental physical or biological laws, even in scenarios where training data is limited or noisy.
Minimizing the ‘ProcessResidual’ serves as a regularization technique within the PINN framework. The ProcessResidual, calculated as the difference between the neural network’s predicted output and the corresponding output from the LINTUL5 biophysical model, quantifies the deviation from established physical constraints. By incorporating the ProcessResidual as a loss term during training, the network is penalized for predictions that significantly diverge from LINTUL5’s established behavior. This constraint effectively guides the learning process, promoting solutions that adhere to known biophysical principles. Consequently, the model requires fewer extensive training datasets to achieve comparable or improved generalization performance, as the biophysical constraint reduces the solution space and mitigates overfitting to potentially noisy or limited data.
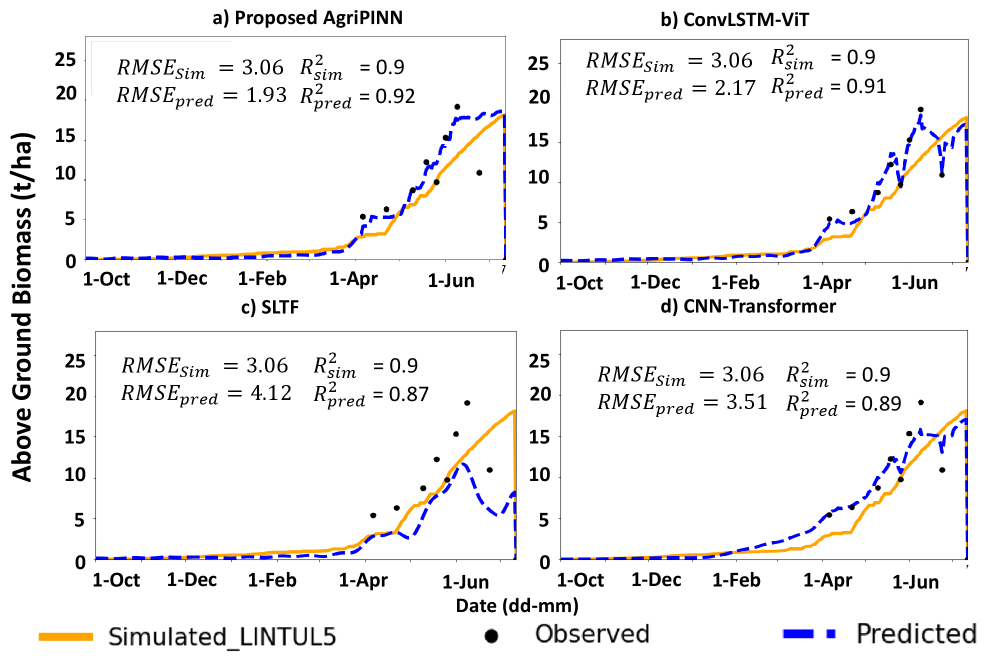
Empirical Validation of Predictive Accuracy
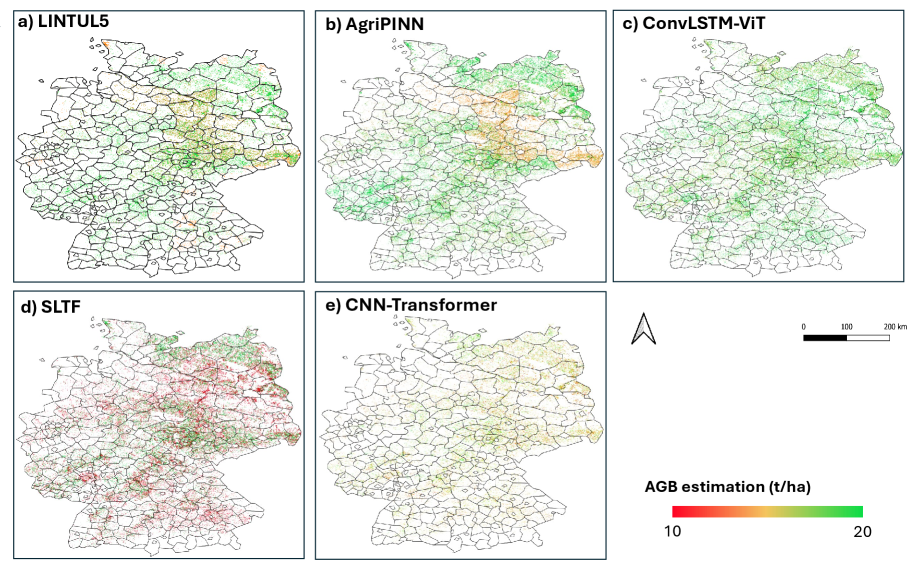
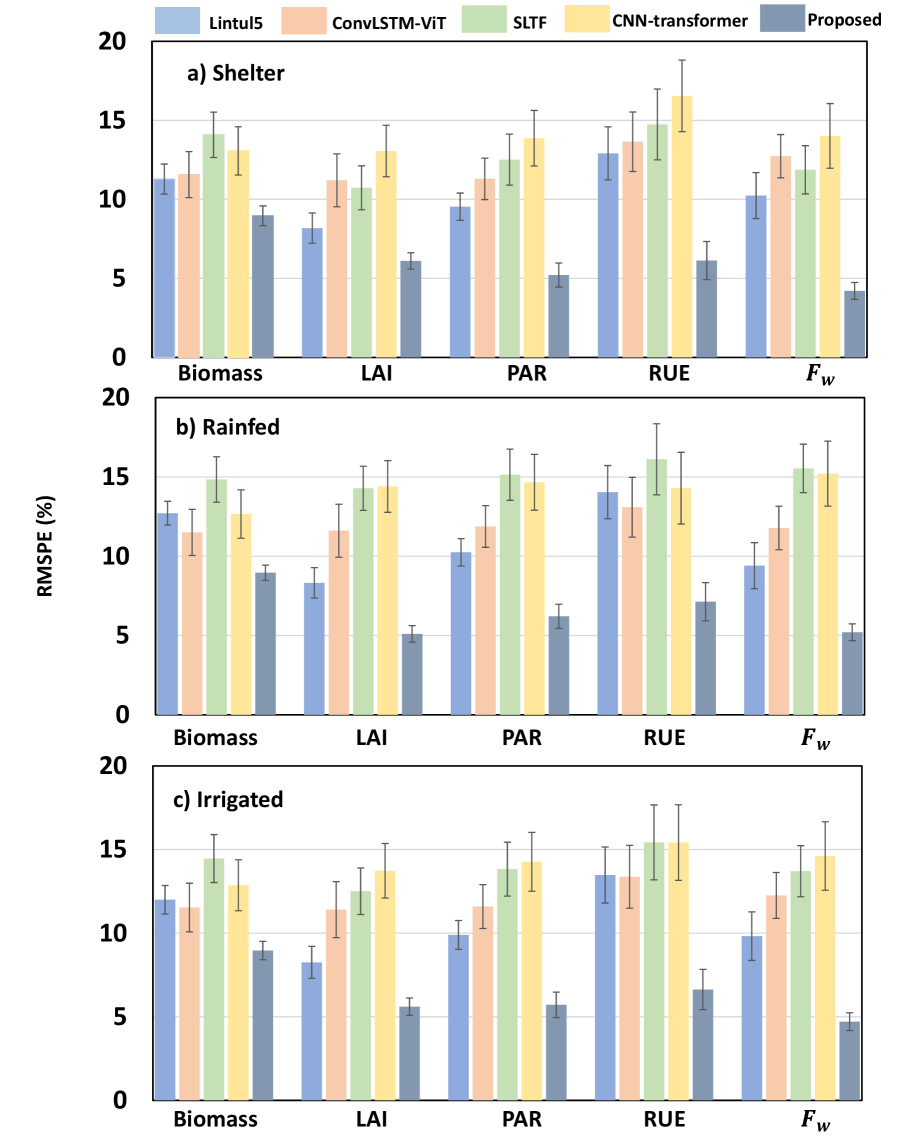
The Physics-Informed Neural Network (PINN) exhibited strong performance in predicting Aboveground Biomass (AGB), as quantified by an R2 value of 0.837. This indicates that 83.7% of the variance in AGB can be explained by the model. Further assessment using the Root Mean Squared Error (RMSE) yielded a value of 2.01, representing the average magnitude of the error between predicted and observed AGB values across a range of environmental conditions and species. These metrics demonstrate the PINN’s ability to accurately estimate AGB, even when applied to datasets not used during training.
The developed framework extends beyond Aboveground Biomass (AGB) prediction to accurately infer critical plant physiological variables. Specifically, the model demonstrates reliable estimation of Leaf Area Index (LAI), a key indicator of vegetation density and photosynthetic capacity; Photosynthetically Active Radiation (PAR), representing the energy available for photosynthesis; and Radiation Use Efficiency (RUE), quantifying the biomass produced per unit of absorbed radiation. Accurate inference of these variables facilitates a more comprehensive understanding of plant function and ecosystem productivity, providing data beyond simple biomass estimation.
The PINN framework’s adaptability was assessed by implementing it with both Convolutional Neural Network (CNN) and Transformer architectures as backbones. Evaluation across various datasets and conditions demonstrated that the predictive performance remained consistent irrespective of the chosen backbone. Specifically, both CNN and Transformer implementations achieved comparable accuracy in estimating Aboveground Biomass (AGB) and other physiological variables, indicating the PINN’s robustness and flexibility in accommodating different neural network structures without significant performance degradation. This suggests the PINN’s core physics-informed loss function effectively guides training regardless of the specific network architecture employed.
Towards a Predictive and Sustainable Agricultural Future
The architecture of this modeling framework prioritizes adaptability through a modular design, heavily reliant on the SIMPLACE infrastructure. This allows for the straightforward incorporation of existing, well-established process-based models – those detailing physiological processes like photosynthesis and nutrient uptake – into the system. Consequently, the framework isn’t limited to a single crop or environment; it can be readily extended to simulate a diverse array of agricultural systems, from temperate cereals to tropical legumes, and across varying climates and soil conditions. This plug-and-play capability drastically reduces the time and resources required to build specialized crop models, fostering broader application and accelerating agricultural research.
The current trajectory of crop modeling stands to be fundamentally altered by a shift towards approaches that demand less extensive data while simultaneously enhancing predictive power. Traditional process-based models, while theoretically sound, often require exhaustive datasets for calibration and validation, limiting their application across diverse agricultural landscapes. This new framework addresses this limitation by intelligently leveraging available data, leading to more accurate simulations even with reduced input. Consequently, agricultural practitioners gain access to tools that facilitate more informed decisions regarding irrigation, fertilization, and pest management, ultimately contributing to increased yields and sustainable farming practices. The potential for widespread adoption lies in the framework’s ability to bridge the gap between data scarcity and the need for precise, reliable crop predictions.
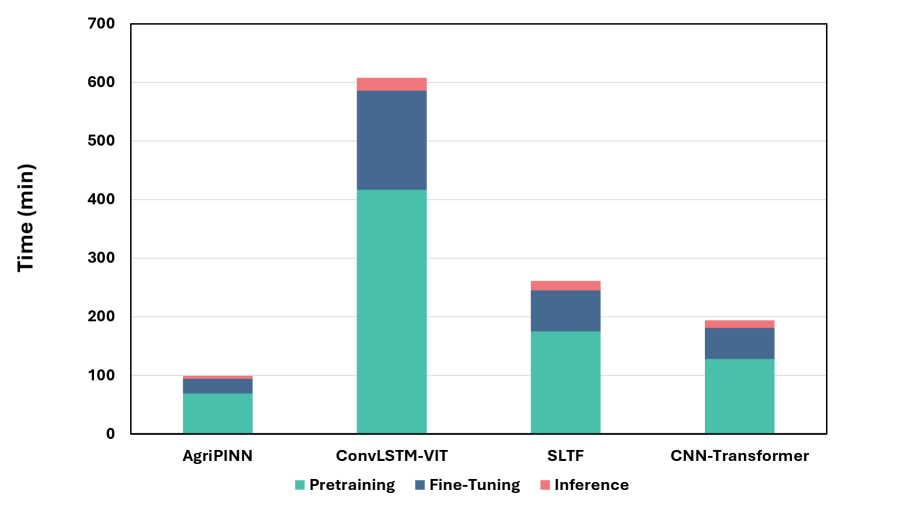
The developed framework achieves a substantial leap in computational efficiency, demonstrably reducing training time by a factor of eight when contrasted with conventional process-based simulations. This acceleration isn’t merely a matter of speed; it’s coupled with a significant decrease in the number of parameters required compared to the most advanced data-driven models currently available. This reduction in complexity not only streamlines the modeling process but also enhances the framework’s robustness and generalizability, allowing it to be applied effectively across diverse agricultural scenarios with fewer input requirements and a lessened risk of overfitting to specific datasets. The combined effect of faster training and reduced parameterization promises a practical and scalable solution for widespread adoption in both research and applied agricultural settings.
Investigations are now directed towards leveraging this framework for dynamic agricultural management. The system is being adapted to ingest real-time data streams – incorporating inputs from sources like remote sensing, weather stations, and field sensors – to provide continuous crop monitoring and early detection of stress factors. This near-instantaneous assessment will enable more accurate yield predictions at various growth stages, moving beyond traditional end-of-season estimates. Critically, the framework’s ability to rapidly simulate different scenarios will allow for the optimization of resource allocation – including water, fertilizer, and pesticides – tailoring inputs to specific crop needs and maximizing efficiency while minimizing environmental impact. This transition promises a shift from reactive agricultural practices to proactive, data-driven strategies.
The pursuit of accurate crop biomass prediction, as demonstrated by AgriPINN, echoes a fundamental tenet of computational rigor. It isn’t simply about achieving a desired output, but about why that output is generated. As Donald Knuth famously stated, “Premature optimization is the root of all evil.” This resonates deeply with the hybrid modeling approach detailed in the paper; AgriPINN doesn’t blindly optimize for accuracy, but rather integrates process-based knowledge – the underlying ‘physics’ of plant growth – to ensure a solution grounded in verifiable principles. This deliberate construction, prioritizing understanding over mere performance, reflects a commitment to elegance and robustness, avoiding the pitfalls of opaque, ‘black box’ predictions. The framework’s interpretability, a key feature, stems from this very adherence to mathematical purity and verifiable logic.
Beyond the Horizon
The presented AgriPINN framework, while a demonstrable advance, merely scratches the surface of a fundamental challenge: the reconciliation of data-driven approximation with demonstrable physical law. Current implementations, elegant as they may be, still rely on empirical validation-a pragmatic concession, but one which ultimately fails the test of mathematical rigor. The asymptotic behavior of these hybrid models, particularly under extreme or novel environmental conditions, remains largely unexplored, and thus, unpredictable. A truly robust solution necessitates a formal proof of convergence and stability, rather than reliance on observational correlation.
Future research must address the limitations inherent in the discretization of continuous biophysical processes. The current reliance on finite difference or finite element approximations introduces error, and a more nuanced approach-perhaps leveraging techniques from fractional calculus or geometric integration-could yield superior results. Furthermore, the generalization capabilities of AgriPINN, while promising, are constrained by the data upon which it is trained. Active learning strategies, coupled with physics-based augmentation, could significantly improve performance in data-scarce environments.
The ultimate goal, however, should not be simply to predict biomass, but to understand the underlying mechanisms governing plant growth under stress. AgriPINN, or its successors, must evolve beyond being a sophisticated regression tool and become a platform for hypothesis testing and the refinement of ecological theory. Only then will such models transcend the realm of empirical utility and achieve true scientific validity.
Original article: https://arxiv.org/pdf/2601.16045.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- YouTuber streams himself 24/7 in total isolation for an entire year
- Gold Rate Forecast
- Lacari banned on Twitch & Kick after accidentally showing explicit files on notepad
- Ragnarok X Next Generation Class Tier List (January 2026)
- Shameless is a Massive Streaming Hit 15 Years Later
- ‘That’s A Very Bad Idea.’ One Way Chris Rock Helped SNL’s Marcello Hernández Before He Filmed His Netflix Special
- We Need to Talk About Will
- Mark Ruffalo Finally Confirms Whether The Hulk Is In Avengers: Doomsday
- Return to Silent Hill Star Breaks Down Her “Daunting” Experience on the Sequel Film
- Ex-Rate My Takeaway star returns with new YouTube channel after “heartbreaking” split
2026-01-23 22:53