Author: Denis Avetisyan
A new deep learning framework leverages multi-modal brain imaging and patient data to enhance the accuracy of Parkinson’s and Alzheimer’s disease diagnosis.
Researchers introduce DW-DGAT, a dynamically weighted dual graph attention network for fusing neuroimaging and phenotypic data to improve neurodegenerative disease diagnosis.
Early and accurate diagnosis of neurodegenerative diseases like Parkinson’s and Alzheimer’s remains a significant clinical challenge due to the complexity and heterogeneity of multi-metric patient data. This paper introduces ‘DW-DGAT: Dynamically Weighted Dual Graph Attention Network for Neurodegenerative Disease Diagnosis’, a novel deep learning framework designed to integrate diverse neuroimaging and phenotypic information via a dual graph attention network. By effectively fusing these data modalities and addressing class imbalance, DW-DGAT achieves state-of-the-art diagnostic performance on benchmark datasets. Could this approach pave the way for more proactive and personalized interventions in the fight against these devastating conditions?
Whispers of Disconnect: Mapping the Brain’s Pre-Symptomatic Shifts
The insidious onset of neurodegenerative diseases, such as Alzheimer’s and Parkinson’s, is increasingly understood as a process that begins decades before noticeable cognitive or motor impairments emerge. Research indicates that the brain’s intricate network of connections – responsible for everything from memory and movement to personality – undergoes subtle but critical alterations long before clinical symptoms manifest. These early changes aren’t necessarily localized to specific brain regions traditionally associated with these diseases; instead, they often appear as disruptions in the communication between different areas. This means that even in the absence of detectable tissue loss or protein aggregation, the brain’s functional architecture is already shifting, foreshadowing the eventual decline. Identifying these pre-symptomatic connectivity changes presents a significant opportunity for early detection and, potentially, the development of interventions designed to slow or even prevent disease progression.
The brain’s capacity for complex thought and action arises not from isolated regions, but from the intricate interplay between them – these are brain regional networks. Precisely mapping these networks is becoming increasingly vital for the early detection of neurodegenerative diseases, as subtle disruptions in connectivity often precede the manifestation of clinical symptoms by years, even decades. Identifying these early alterations allows for potential intervention strategies aimed at slowing disease progression or, ideally, preventing symptom onset. The ability to visualize and quantify the strength and efficiency of connections between different brain areas offers a biomarker – a measurable indicator – of underlying pathological processes, opening new avenues for personalized medicine and proactive healthcare focused on preserving cognitive function.
Conventional neuroimaging analyses frequently treat brain regions as isolated entities, failing to fully appreciate the intricate web of connections that define healthy brain function. This simplification limits the detection of subtle disruptions indicative of early neurodegenerative disease; the brain doesn’t fail in isolated pockets, but rather through a cascading breakdown of interconnected networks. Traditional methods often rely on averaging signals across large regions, obscuring the nuanced, individual-level changes in connectivity that precede noticeable symptoms. Consequently, these techniques lack the sensitivity required to pinpoint the earliest signs of disease, hindering preventative interventions and delaying effective treatment strategies. A more holistic approach, capable of mapping the dynamic interplay between brain regions, is therefore essential for improving early detection rates and ultimately, patient outcomes.
The advent of large-scale longitudinal studies, prominently including the Alzheimer’s Disease Neuroimaging Initiative 3 (ADNI3) and the Parkinson’s Progression Markers Initiative (PPMI), has revolutionized the potential for early disease detection, yet simultaneously presents considerable analytical challenges. These ongoing investigations amass extensive datasets of neuroimaging information – encompassing structural MRI, functional MRI, and PET scans – collected from participants over many years. The sheer volume and complexity of this data necessitate the development of advanced computational tools, moving beyond traditional statistical methods. Sophisticated techniques, such as graph theory, machine learning algorithms, and network-based statistics, are now crucial for effectively processing this wealth of information, discerning subtle shifts in brain connectivity, and ultimately identifying biomarkers that signal disease onset long before symptoms manifest clinically. The continued refinement of these analytical approaches promises to unlock the full potential of longitudinal neuroimaging data in the fight against neurodegenerative diseases.
Weaving the Map: Data Fusion and Graph Construction
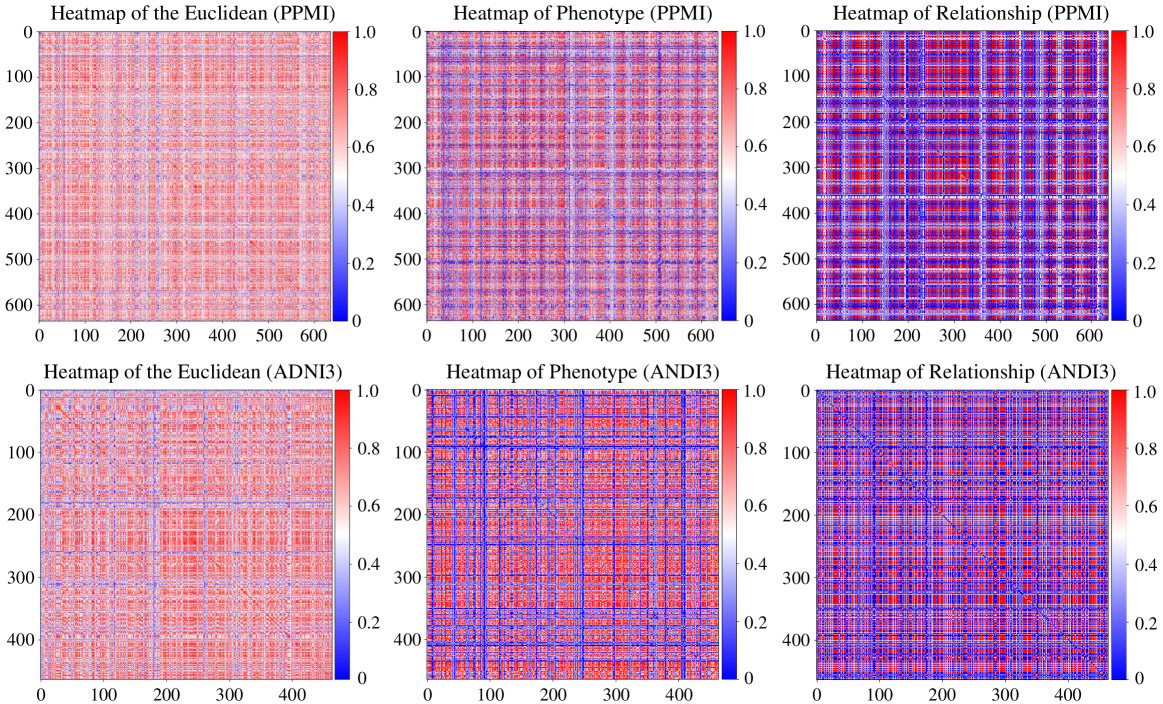
The integration of data from multiple neuroimaging modalities, specifically Magnetic Resonance Imaging (MRI) and Diffusion Tensor Imaging (DTI), significantly enhances the characterization of brain organization. MRI provides high-resolution anatomical images detailing brain structure, while DTI assesses the white matter tracts that facilitate communication between brain regions. MRI data reveals macroscopic anatomical features and identifies structural abnormalities, whereas DTI measures the direction and strength of water diffusion along axonal fibers, indicating the integrity and organization of white matter pathways. Combining these datasets allows for a more complete understanding of both the brain’s physical structure and its functional connectivity, enabling researchers to investigate relationships between anatomical features, white matter integrity, and cognitive performance.
Data fusion techniques are critical for combining MRI and Diffusion Tensor Imaging (DTI) data due to the inherent differences in their acquisition parameters and data representations. Successful integration necessitates precise spatial alignment, often achieved through image registration algorithms, to ensure corresponding anatomical locations are identified across modalities. Furthermore, differing noise characteristics and signal intensities require normalization and standardization procedures. Variations in spatial resolution between MRI and DTI necessitate resampling techniques to establish a common space for analysis. Incorrect or insufficient alignment and processing can introduce artifacts and inaccuracies, compromising the validity of subsequent analyses and interpretations derived from the combined dataset.
The PANDA pipeline is employed for preprocessing of both Magnetic Resonance Imaging (MRI) and Diffusion Tensor Imaging (DTI) data to maintain data integrity and consistency. This pipeline incorporates steps for bias field correction, skull stripping, spatial normalization to a standard template – such as MNI space – and segmentation of brain tissues. For DTI data, PANDA includes processing for eddy current correction and diffusion tensor estimation. These standardized procedures minimize noise and artifacts, and ensure comparability across subjects, which is crucial for downstream analyses involving data fusion and graph construction. The pipeline’s robustness is validated through established quality control metrics and automated checks for common preprocessing errors.
The construction of a brain graph from fused MRI and DTI data involves defining discrete brain regions as nodes, typically based on anatomical atlases or functional parcellation schemes. Edges connecting these nodes represent structural or functional relationships, with edge weight quantifying the strength of connection. These weights are derived from metrics calculated from the fused data; for example, fractional anisotropy (FA) from DTI can indicate the integrity of white matter tracts and contribute to edge weight, while functional connectivity derived from MRI data can indicate statistical dependence between regional activity. The resulting graph provides a network-based representation of brain organization, facilitating the application of graph theory metrics to characterize brain connectivity and identify key network properties.
Dual-Scale Perception: Capturing Connectivity with Attention Networks
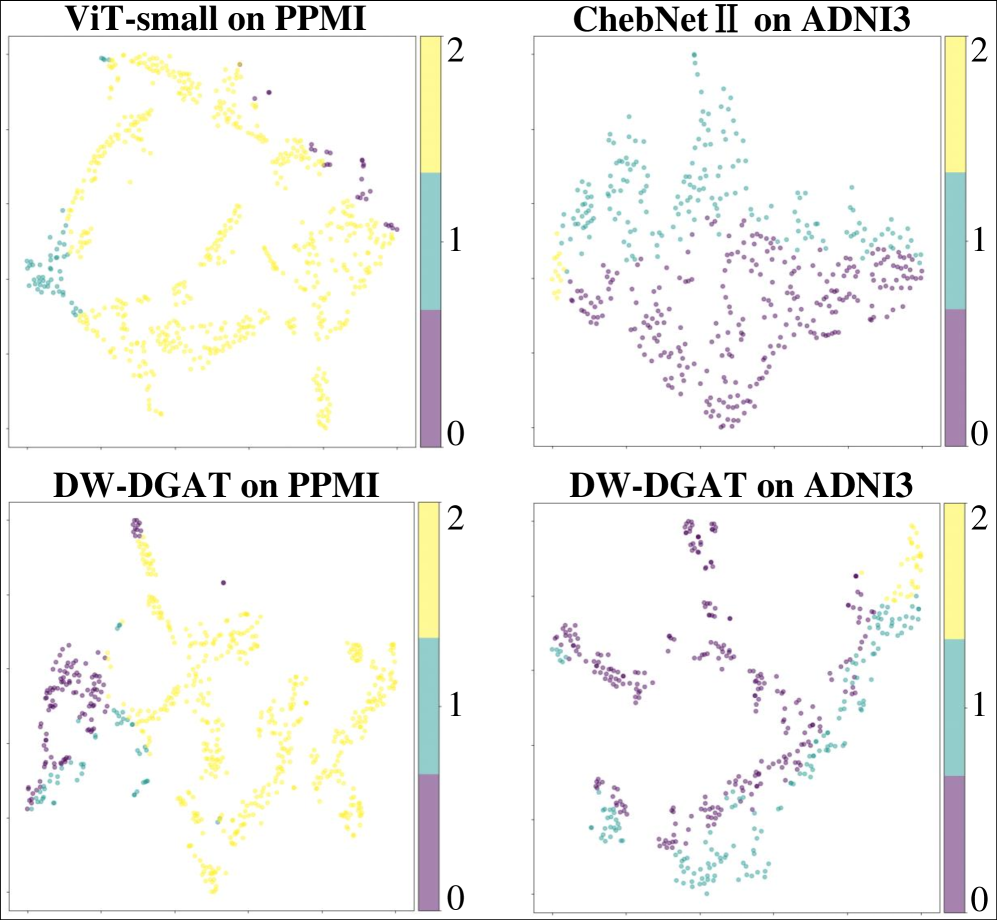
The Dual Graph Attention Network is designed to analyze brain connectivity at multiple scales. It accomplishes this by leveraging two distinct graph attention modules: a Single Graph Attention (SGA) module focuses on localized feature extraction from individual regions of interest (ROIs), effectively capturing micro-level connectivity patterns. Simultaneously, a Global Graph Attention (GGA) module operates on the entire brain network to model long-range dependencies and macro-level interactions between ROIs. This dual-scale approach allows the network to represent both fine-grained, localized brain activity and broad, network-wide connectivity patterns, providing a more comprehensive representation of brain structure and function.
The Dual Graph Attention Network employs two distinct graph attention modules to analyze brain connectivity at multiple scales. The Single Graph Attention (SGA) module processes connectivity data derived from individual regions of interest (ROIs), extracting localized features specific to each brain area. Complementing this, the Global Graph Attention (GGA) module operates on the entire brain network, identifying broader relationships and dependencies between ROIs that may not be apparent when considering them in isolation. This dual-module approach allows the network to capture both fine-grained, local connectivity patterns and large-scale, holistic brain network organization, contributing to a more comprehensive representation of brain structure and function.
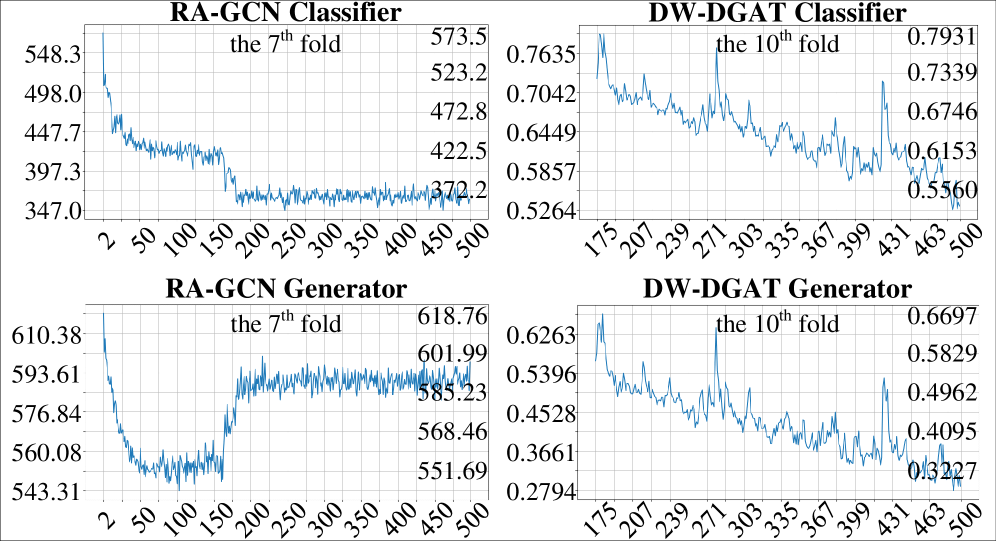
The Class Weight Generator (CWG) addresses the prevalent issue of class imbalance in medical datasets by dynamically adjusting the weighting assigned to each class during model training. Traditional machine learning approaches often treat all classes equally, which can lead to biased models that perform poorly on minority classes – a common scenario in neurological disease classification where the number of healthy controls typically exceeds the number of patients with a specific condition. The CWG calculates weights inversely proportional to the class frequency within each training batch, effectively increasing the contribution of under-represented classes to the loss function and preventing the model from being dominated by the majority class. This batch-wise adjustment ensures that the model receives a more balanced learning signal, improving its ability to accurately identify and classify all conditions represented in the dataset.
The Dual Graph Attention Network demonstrates state-of-the-art performance in analyzing brain connectivity data, achieving 92% accuracy on both the Alzheimer’s Disease Neuroimaging Initiative 3 (ADNI3) and Parkinson’s Progression Markers Initiative (PPMI) datasets. Comparative analysis reveals a significant performance advantage over the next best performing network, with improvements of 7.57% on the PPMI dataset and 4.62% on the ADNI3 dataset. These results indicate the network’s ability to effectively model complex brain connectivity patterns and improve diagnostic accuracy in neurological conditions.
Whispers Become Warnings: DTI Metrics as Early Biomarkers
Investigations into white matter integrity reveal that subtle alterations, often preceding observable clinical symptoms, can be effectively detected through Diffusion Tensor Imaging (DTI). Analyses demonstrate the particular sensitivity of several DTI-derived metrics – including Fractional Anisotropy (FA), which measures the directionality of water diffusion; Mean Diffusivity (MD), reflecting the overall magnitude of diffusion; Axial Diffusivity (AXD), characterizing diffusion along nerve fibers; Radial Diffusivity (RDD), measuring diffusion perpendicular to fibers; and Local Diffusion Homogeneity (LDH), indicating the consistency of diffusion within a tissue – to these early changes. These metrics collectively provide a nuanced understanding of white matter structure, allowing for the identification of disruptions in connectivity before widespread neurodegeneration occurs and offering potential for proactive intervention strategies.
Diffusion Tensor Imaging (DTI) metrics offer a powerful means of characterizing white matter integrity, and crucially, can reveal alterations in brain connectivity long before the emergence of noticeable clinical symptoms. Subtle changes in these metrics – including Fractional Anisotropy, Mean Diffusivity, and others – reflect early disruptions to the organized structure of white matter tracts, which are vital for efficient communication between brain regions. These quantifiable measures act as biomarkers, signaling a potential vulnerability or the very earliest stages of neurodegenerative processes. By detecting these subtle shifts, researchers gain the opportunity to track disease progression at a pre-symptomatic level, potentially paving the way for targeted interventions designed to slow or even prevent the onset of debilitating conditions.
The application of a novel graph-based deep learning framework, incorporating diffusion tensor imaging (DTI)-derived biomarkers, demonstrates a high degree of accuracy in distinguishing individuals at risk for neurodegenerative disease. Evaluation on two prominent datasets – the Parkinson’s Progression Markers Initiative (PPMI) and the Alzheimer’s Disease Neuroimaging Initiative 3 (ADNI3) – yielded balanced accuracies of 79.43% and 77.58%, respectively. These results suggest the framework effectively leverages subtle changes in white matter integrity, as quantified by metrics like Fractional Anisotropy and Mean Diffusivity, to identify patterns indicative of early-stage pathology. This performance underscores the potential for utilizing these biomarkers within a deep learning architecture to improve diagnostic capabilities and facilitate timely interventions.
The potential for earlier intervention in neurodegenerative diseases represents a significant advancement facilitated by this research. Detecting subtle changes in brain connectivity before the onset of overt clinical symptoms allows for proactive strategies, potentially slowing disease progression and preserving cognitive function. This approach moves beyond reactive treatment, offering the possibility of neuroprotective interventions tailored to the individual’s unique brain connectivity profile. Improved patient outcomes are anticipated not only through earlier diagnosis, but also by enabling clinicians to monitor treatment efficacy with greater precision, optimizing therapeutic regimens and enhancing the quality of life for those affected by these debilitating conditions. The ability to track subtle shifts in white matter integrity serves as a crucial step toward personalized medicine in the fight against neurodegeneration.
The pursuit of diagnostic precision, as outlined in this work concerning DW-DGAT and neurodegenerative diseases, feels less like engineering and more like coaxing a fickle spirit. The model doesn’t simply learn; it negotiates with the chaos inherent in multi-dimensional data. It’s a delicate dance – fusing imaging with phenotypic information, attempting to discern signal from the inevitable noise. As Fei-Fei Li once observed, “Data isn’t numbers – it’s whispers of chaos.” This sentiment perfectly encapsulates the challenge; the network doesn’t conquer the complexity, it persuades it to reveal its secrets, building a fragile spell against the encroaching darkness of diseases like Parkinson’s and Alzheimer’s. Each layer, each attention mechanism, is a carefully chosen incantation, hoping to hold back the tide.
What Lies Ahead?
The construction of DW-DGAT, as with any attempt to coax order from the entropy of biological systems, feels less like a solution and more like a beautifully arranged postponement. The network skillfully marries imaging data with phenotypic whispers, but the true signal – the unfolding of neurodegeneration – remains stubbornly beyond complete capture. Any correlation achieved is, naturally, suspect; a testament not to insight, but to the limitations of the measurements themselves. If the hypothesis held, one must question whether the inquiry truly reached the core of the problem.
Future iterations will inevitably seek greater dimensional integration – genomic data, proteomic signatures, perhaps even the elusive quantification of lived experience. Yet, the challenge isn’t simply accumulation; it’s discerning what data deserves attention. The signal, after all, is always drowned in noise. More sophisticated graph structures might emerge, mimicking the brain’s complexity with ever-finer detail, but the model will remain a simplification, a map mistaken for the territory.
The ultimate test isn’t diagnostic accuracy, but predictive power. Can these networks anticipate decline before clinical manifestation? That would be a truly unsettling achievement – not because it solves a medical problem, but because it reveals just how predetermined our fates might be. And anything easily predicted, anything cleanly measurable, is almost certainly not worth knowing.
Original article: https://arxiv.org/pdf/2601.10001.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Gold Rate Forecast
- Pokemon Legends: Z-A Is Giving Away A Very Big Charizard
- Six Flags Qiddiya City Closes Park for One Day Shortly After Opening
- How to Complete the Behemoth Guardian Project in Infinity Nikki
- 10 Worst Sci-Fi Movies of All Time, According to Richard Roeper
- Bitcoin After Dark: The ETF That’s Sneakier Than Your Ex’s Texts at 2AM 😏
- Dev Plans To Voluntarily Delete AI-Generated Game
- Fans pay respects after beloved VTuber Illy dies of cystic fibrosis
- Stephen King Is Dominating Streaming, And It Won’t Be The Last Time In 2026
- Stranger Things Season 5 & ChatGPT: The Truth Revealed
2026-01-19 06:11